Population genetics in microchannels
- PMID: 35302890
- PMCID: PMC8944277
- DOI: 10.1073/pnas.2120821119
Population genetics in microchannels
Abstract
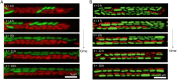
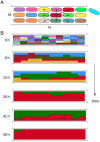
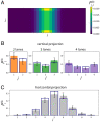
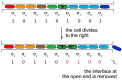
SignificanceMany microbial populations proliferate in small channels. In such environments, reproducing cells organize in parallel lanes. Reproducing cells shift these lanes, potentially expelling other cells from the channel. In this paper, we combine theory and experiments to understand how these dynamics affects the diversity of a microbial population. We theoretically predict that genetic diversity is quickly lost along lanes of cells. Our experiments confirm that a population of proliferating Escherichia coli in a microchannel organizes into lanes of genetically identical cells within a few generations. Our findings elucidate the effect of lane formation on populations evolution, with potential applications ranging from microbial ecology in soil to dynamics of epithelial tissues in higher organisms.
Keywords: bacterial evolution; individual-based models; microfluidics; spatial population dynamics.
Conflict of interest statement
The authors declare no competing interest.
Figures


References
-
- You Z., Pearce D. J., Sengupta A., Giomi L., Geometry and mechanics of microdomains in growing bacterial colonies. Phys. Rev. X 8, 031065 (2018).
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

