Mucus sialylation determines intestinal host-commensal homeostasis
- PMID: 35303419
- PMCID: PMC9088855
- DOI: 10.1016/j.cell.2022.02.013
Mucus sialylation determines intestinal host-commensal homeostasis
Abstract
Intestinal mucus forms the first line of defense against bacterial invasion while providing nutrition to support microbial symbiosis. How the host controls mucus barrier integrity and commensalism is unclear. We show that terminal sialylation of glycans on intestinal mucus by ST6GALNAC1 (ST6), the dominant sialyltransferase specifically expressed in goblet cells and induced by microbial pathogen-associated molecular patterns, is essential for mucus integrity and protecting against excessive bacterial proteolytic degradation. Glycoproteomic profiling and biochemical analysis of ST6 mutations identified in patients show that decreased sialylation causes defective mucus proteins and congenital inflammatory bowel disease (IBD). Mice harboring a patient ST6 mutation have compromised mucus barriers, dysbiosis, and susceptibility to intestinal inflammation. Based on our understanding of the ST6 regulatory network, we show that treatment with sialylated mucin or a Foxo3 inhibitor can ameliorate IBD.
Keywords: ST6GalNAc1; dysbiosis; glycobiology; human genetic disease; inflammatory bowel disease; intestinal homeostasis; intestinal stem cells; mucus barrier; short-chain fatty acids; sialylation.
Copyright © 2022. Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
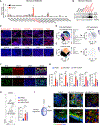



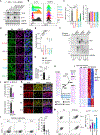

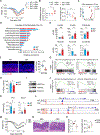
Comment in
-
Mucus Sialylation Maintains the Peace in Intestinal Host Microbe Relations.Gastroenterology. 2022 Aug;163(2):527-528. doi: 10.1053/j.gastro.2022.05.041. Epub 2022 Jun 1. Gastroenterology. 2022. PMID: 35661726 No abstract available.
References
-
- Alipour M, Zaidi D, Valcheva R, Jovel J, Martinez I, Sergi C, Walter J, Mason AL, Wong GK, Dieleman LA, et al. (2016). Mucosal Barrier Depletion and Loss of Bacterial Diversity are Primary Abnormalities in Paediatric Ulcerative Colitis. Journal of Crohn’s & colitis 10, 462–471. 10.1093/ecco-jcc/jjv223. - DOI - PMC - PubMed
-
- Banerjee A, Herring CA, Chen B, Kim H, Simmons AJ, Southard-Smith AN, Allaman MM, White JR, Macedonia MC, McKinley ET, et al. (2020). Succinate Produced by Intestinal Microbes Promotes Specification of Tuft Cells to Suppress Ileal Inflammation. Gastroenterology 159, 2101–2115 e2105. 10.1053/j.gastro.2020.08.029. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

