Cholesterol promotes EGFR-TKIs resistance in NSCLC by inducing EGFR/Src/Erk/SP1 signaling-mediated ERRα re-expression
- PMID: 35303882
- PMCID: PMC8932110
- DOI: 10.1186/s12943-022-01547-3
Cholesterol promotes EGFR-TKIs resistance in NSCLC by inducing EGFR/Src/Erk/SP1 signaling-mediated ERRα re-expression
Abstract
Background: The use of epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs) brings remarkable benefits for the survival of patients with advanced NSCLC harboring EGFR mutations. Unfortunately, acquired resistance seems to be inevitable and limits the application of EGFR-TKIs in clinical practice. This study reported a common molecular mechanism sustaining resistance and potential treatment options to overcome EGFR-TKIs resistance.
Methods: EGFR-TKIs resistant NSCLC cells were established and confirmed by MTT assay. Cholesterol content was detected and the promotional function of cholesterol on NSCLC growth was determined in vivo. Then, we identified ERRα expression as the downstream factor of cholesterol-mediated drug resistance. To dissect the regulatory mechanism, we conducted experiments, including immunofluorescence, co-immunoprecipitation, luciferase reporter assay and chromatin immunoprecipitation assay.
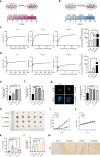
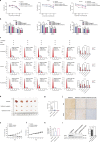
Results: Long-term exposure to EGFR-TKIs generate drug resistance with the characteristic of cholesterol accumulation in lipid rafts, which promotes EGFR and Src to interact and lead EGFR/Src/Erk signaling reactivation-mediated SP1 nuclear translocation and ERRα re-expression. Further investigation identifies ERRα as a target gene of SP1. Functionally, re-expression of ERRα sustains cell proliferation by regulating ROS detoxification process. Lovastatin, a drug used to decrease cholesterol level, and XCT790, an inverse agonist of ERRα, overcome gefitinib and osimertinib resistance both in vitro and in vivo.
Conclusions: Our study indicates that cholesterol/EGFR/Src/Erk/SP1 axis-induced ERRα re-expression promotes survival of gefitinib and osimertinib-resistant cancer cells. Besides, we demonstrate the potential of lowing cholesterol and downregulation of ERRα as effective adjuvant treatment of NSCLC.
Keywords: Cholesterol; EGFR-TKIs resistance; Non-small cell lung cancer.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
-
- Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352(8):786–792. - PubMed
-
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. - PubMed
-
- Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361(10):947–957. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

