A cationic lipid mediated CRISPR/Cas9 technique for the production of stable genome edited citrus plants
- PMID: 35303912
- PMCID: PMC8932238
- DOI: 10.1186/s13007-022-00870-6
A cationic lipid mediated CRISPR/Cas9 technique for the production of stable genome edited citrus plants
Abstract
Background: The genetic engineering of crops has enhanced productivity in the face of climate change and a growing global population by conferring desirable genetic traits, including the enhancement of biotic and abiotic stress tolerance, to improve agriculture. The clustered regularly interspaced short palindromic repeats (CRISPR/Cas9) system has been found to be a promising technology for genomic editing. Protoplasts are often utilized for the development of genetically modified plants through in vitro integration of a recombinant DNA fragment into the plant genome. We targeted the citrus Nonexpressor of Pathogenesis-Related 3 (CsNPR3) gene, a negative regulator of systemic acquired resistance (SAR) that governs the proteasome-mediated degradation of NPR1 and developed a genome editing technique targeting citrus protoplast DNA to produce stable genome-edited citrus plants.
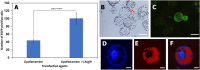
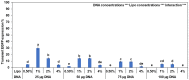
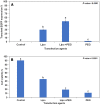
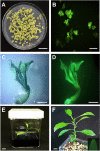
Results: Here, we determined the best cationic lipid nanoparticles to deliver donor DNA and described a protocol using Lipofectamine™ LTX Reagent with PLUS Reagent to mediate DNA delivery into citrus protoplasts. A Cas9 construct containing a gRNA targeting the CsNPR3 gene was transfected into citrus protoplasts using the cationic lipid transfection agent Lipofectamine with or without polyethylene glycol (PEG, MW 6000). The optimal transfection efficiency for the encapsulation was 30% in Lipofectamine, 51% in Lipofectamine with PEG, and 2% with PEG only. Additionally, plasmid encapsulation in Lipofectamine resulted in the highest cell viability percentage (45%) compared with PEG. Nine edited plants were obtained and identified based on the T7EI assay and Sanger sequencing. The developed edited lines exhibited downregulation of CsNPR3 expression and upregulation of CsPR1.
Conclusions: Our results demonstrate that utilization of the cationic lipid-based transfection agent Lipofectamine is a viable option for the successful delivery of donor DNA and subsequent successful genome editing in citrus.
Keywords: CRISPR/Cas9; Citrus; Genome editing; Lipofection; NPR3; Protoplast; Systemic acquired resistance (SAR).
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
CRISPR technology towards genome editing of the perennial and semi-perennial crops citrus, coffee and sugarcane.Front Plant Sci. 2024 Jan 8;14:1331258. doi: 10.3389/fpls.2023.1331258. eCollection 2023. Front Plant Sci. 2024. PMID: 38259920 Free PMC article. Review.
-
Cationic lipid nanoparticle-mediated delivery of a Cas9/crRNA ribonucleoprotein complex for transgene-free editing of the citrus plant genome.Plant Cell Rep. 2024 Jun 14;43(7):171. doi: 10.1007/s00299-024-03254-3. Plant Cell Rep. 2024. PMID: 38874819
-
Genetic and physiological characteristics of CsNPR3 edited citrus and their impact on HLB tolerance.Front Genome Ed. 2024 Dec 4;6:1485529. doi: 10.3389/fgeed.2024.1485529. eCollection 2024. Front Genome Ed. 2024. PMID: 39698041 Free PMC article.
-
An efficient DNA- and selectable-marker-free genome-editing system using zygotes in rice.Nat Plants. 2019 Apr;5(4):363-368. doi: 10.1038/s41477-019-0386-z. Epub 2019 Mar 25. Nat Plants. 2019. PMID: 30911123
-
Highly Efficient Genome Editing in Plant Protoplasts by Ribonucleoprotein Delivery of CRISPR-Cas12a Nucleases.Front Genome Ed. 2022 Jan 31;4:780238. doi: 10.3389/fgeed.2022.780238. eCollection 2022. Front Genome Ed. 2022. PMID: 35174354 Free PMC article. Review.
Cited by
-
Cas12a RNP-mediated co-transformation enables transgene-free multiplex genome editing, long deletions, and inversions in citrus chromosome.Front Plant Sci. 2024 Aug 1;15:1448807. doi: 10.3389/fpls.2024.1448807. eCollection 2024. Front Plant Sci. 2024. PMID: 39148610 Free PMC article.
-
Generation of transgene-free canker-resistant Citrus sinensis cv. Hamlin in the T0 generation through Cas12a/CBE co-editing.Front Plant Sci. 2024 Mar 26;15:1385768. doi: 10.3389/fpls.2024.1385768. eCollection 2024. Front Plant Sci. 2024. PMID: 38595767 Free PMC article.
-
CRISPR technology towards genome editing of the perennial and semi-perennial crops citrus, coffee and sugarcane.Front Plant Sci. 2024 Jan 8;14:1331258. doi: 10.3389/fpls.2023.1331258. eCollection 2023. Front Plant Sci. 2024. PMID: 38259920 Free PMC article. Review.
-
Plant Virus-Derived Vectors for Plant Genome Engineering.Viruses. 2023 Feb 14;15(2):531. doi: 10.3390/v15020531. Viruses. 2023. PMID: 36851743 Free PMC article. Review.
-
In planta genome editing in citrus facilitated by co-expression of CRISPR/Cas and developmental regulators.Plant J. 2025 Apr;122(2):e70155. doi: 10.1111/tpj.70155. Plant J. 2025. PMID: 40275470 Free PMC article.
References
-
- Duan YX, Liu X, Fan J, Li DL, Wu RC, Guo WW. Multiple shoot induction from seedling epicotyls and transgenic citrus plant regeneration containing the green fluorescent protein gene. Bot Stud. 2007;48:165–171.
-
- Zhang X, Francis MI, Dawson WO, Graham JH, Orbović V, Triplett EW, Mou Z. Over-expression of the Arabidopsis NPR1 gene in citrus increases resistance to citrus canker. Eur J Plant Pathol. 2010;128:91–100.
-
- Zhang H-Z, Cai XZ. Nonexpressor of pathogenesis-related genes 1 (NPR1): a key node of plant disease resistance signalling network. Chin J Biotechnol. 2005;21:511–515. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

