Comparative analysis of Streptococcus suis genomes identifies novel candidate virulence-associated genes in North American isolates
- PMID: 35303917
- PMCID: PMC8932342
- DOI: 10.1186/s13567-022-01039-8
Comparative analysis of Streptococcus suis genomes identifies novel candidate virulence-associated genes in North American isolates
Abstract
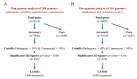
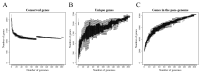
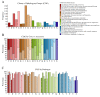
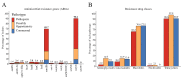
Streptococcus suis is a significant economic and welfare concern in the swine industry. Pan-genome analysis provides an in-silico approach for the discovery of genes involved in pathogenesis in bacterial pathogens. In this study, we performed pan-genome analysis of 208 S. suis isolates classified into the pathogenic, possibly opportunistic, and commensal pathotypes to identify novel candidate virulence-associated genes (VAGs) of S. suis. Using chi-square tests and LASSO regression models, three accessory pan-genes corresponding to S. suis strain P1/7 markers SSU_RS09525, SSU_RS09155, and SSU_RS03100 (>95% identity) were identified as having a significant association with the pathogenic pathotype. The proposed novel SSU_RS09525 + /SSU_RS09155 + /SSU_RS03100 + genotype identified 96% of the pathogenic pathotype strains, suggesting a novel genotyping scheme for predicting the pathogenicity of S. suis isolates in North America. In addition, mobile genetic elements carrying antimicrobial resistance genes (ARGs) and VAGs were identified but did not appear to play a major role in the spread of ARGs and VAGs.
Keywords: Streptococcus suis; comparative genomics; pathotype; virulence-associated genes (VAGs).
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Gottschalk M, Segura M. Streptococcosis. In: Zimmerman JJ, Karriker LA, Ramirez A, Schwartz KJ, Stevenson GW, Zhang J, editors. Diseases of Swine. 11. Hoboken: Wiley; 2019. pp. 934–950.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

