Quantitative glycoproteomics analysis identifies novel FUT8 targets and signaling networks critical for breast cancer cell invasiveness
- PMID: 35303925
- PMCID: PMC8932202
- DOI: 10.1186/s13058-022-01513-3
Quantitative glycoproteomics analysis identifies novel FUT8 targets and signaling networks critical for breast cancer cell invasiveness
Abstract
Background: We recently showed that fucosyltransferase 8 (FUT8)-mediated core fucosylation of transforming growth factor-β receptor enhances its signaling and promotes breast cancer invasion and metastasis. However, the complete FUT8 target glycoproteins and their downstream signaling networks critical for breast cancer progression remain largely unknown.
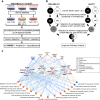
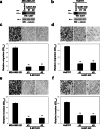
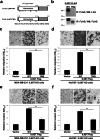
Method: We performed quantitative glycoproteomics with two highly invasive breast cancer cell lines to unravel a comprehensive list of core-fucosylated glycoproteins by comparison to parental wild-type and FUT8-knockout counterpart cells. In addition, ingenuity pathway analysis (IPA) was performed to highlight the most enriched biological functions and signaling pathways mediated by FUT8 targets. Novel FUT8 target glycoproteins with biological interest were functionally studied and validated by using LCA (Lens culinaris agglutinin) blotting and LC-MS/MS (liquid chromatography-tandem mass spectrometry) analysis.
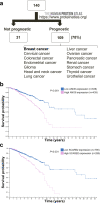
Results: Loss-of-function studies demonstrated that FUT8 knockout suppressed the invasiveness of highly aggressive breast carcinoma cells. Quantitative glycoproteomics identified 140 common target glycoproteins. Ingenuity pathway analysis (IPA) of these target proteins gave a global and novel perspective on signaling networks essential for breast cancer cell migration and invasion. In addition, we showed that core fucosylation of integrin αvβ5 or IL6ST might be crucial for breast cancer cell adhesion to vitronectin or enhanced cellular signaling to interleukin 6 and oncostatin M, two cytokines implicated in the breast cancer epithelial-mesenchymal transition and metastasis.
Conclusions: Our report reveals a comprehensive list of core-fucosylated target proteins and provides novel insights into signaling networks crucial for breast cancer progression. These findings will assist in deciphering the complex molecular mechanisms and developing diagnostic or therapeutic approaches targeting these signaling pathways in breast cancer metastasis.
Keywords: Breast cancer; Core fucosylation; FUT8; Glycoproteomics; Metastasis.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Weigelt B, Peterse JL, van Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer. 2005;5:591–602. - PubMed
-
- Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126:855–867. - PubMed
-
- Miyoshi E, Moriwaki K, Nakagawa T. Biological function of fucosylation in cancer biology. J Biochem. 2008;143:725–729. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

