BRD4 inhibitor GNE987 exerts anti-cancer effects by targeting super-enhancers in neuroblastoma
- PMID: 35303940
- PMCID: PMC8932231
- DOI: 10.1186/s13578-022-00769-8
BRD4 inhibitor GNE987 exerts anti-cancer effects by targeting super-enhancers in neuroblastoma
Abstract
Background: Neuroblastoma (NB) is a common extracranial malignancy with high mortality in children. Recently, super-enhancers (SEs) have been reported to play a critical role in the tumorigenesis and development of NB via regulating a wide range of oncogenes Thus, the synthesis and identification of chemical inhibitors specifically targeting SEs are of great urgency for the clinical therapy of NB. This study aimed to characterize the activity of the SEs inhibitor GNE987, which targets BRD4, in NB.
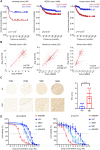
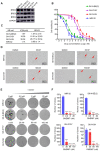
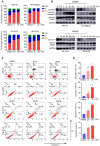
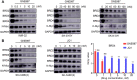
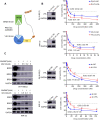
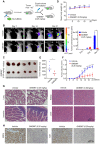
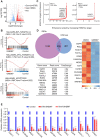
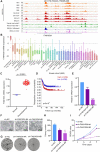
Results: In this study, we found that nanomolar concentrations of GNE987 markedly diminished NB cell proliferation and survival via degrading BRD4. Meanwhile, GNE987 significantly induced NB cell apoptosis and cell cycle arrest. Consistent with in vitro results, GNE987 administration (0.25 mg/kg) markedly decreased the tumor size in the xenograft model, with less toxicity, and induced similar BRD4 protein degradation to that observed in vitro. Mechanically, GNE987 led to significant downregulation of hallmark genes associated with MYC and the global disruption of the SEs landscape in NB cells. Moreover, a novel candidate oncogenic transcript, FAM163A, was identified through analysis of the RNA-seq and ChIP-seq data. FAM163A is abnormally transcribed by SEs, playing an important role in NB occurrence and development.
Conclusion: GNE987 destroyed the abnormal transcriptional regulation of oncogenes in NB by downregulating BRD4, which could be a potential therapeutic candidate for NB.
Keywords: BRD4; Broad H3K27ac domain; Neuroblastoma; PROTAC; Super-enhancer.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
References
-
- Nakagawara A, Li Y, Izumi H, Muramori K, Inada H, Nishi M. Neuroblastoma. Jpn J Clin Oncol. 2018;48(3):214–241. - PubMed
-
- Whittle SB, Smith V, Doherty E, Zhao S, McCarty S, Zage PE. Overview and recent advances in the treatment of neuroblastoma. Expert Rev Anticancer Ther. 2017;17(4):369–386. - PubMed
-
- Wienke J, Dierselhuis MP, Tytgat GAM, Kunkele A, Nierkens S, Molenaar JJ. The immune landscape of neuroblastoma: challenges and opportunities for novel therapeutic strategies in pediatric oncology. Eur J Cancer. 2021;144:123–150. - PubMed
-
- Moreno L, Guo D, Irwin MS, Berthold F, Hogarty M, Kamijo T, Morgenstern D, Pasqualini C, Ash S, Potschger U, et al. A nomogram of clinical and biologic factors to predict survival in children newly diagnosed with high-risk neuroblastoma: an international neuroblastoma risk group project. Pediatr Blood Cancer. 2021;68(3):e28794. - PubMed
Grants and funding
- 81971867/National Natural Science Foundation of China
- 81970163/National Natural Science Foundation of China
- 81802499/National Natural Science Foundation of China
- 81872845/National Natural Science Foundation of China
- 81902534/National Natural Science Foundation of China
- 82072767/National Natural Science Foundation of China
- 52003183/National Natural Science Foundation of China
- SBK2019021442/Natural Science Foundation of Jiangsu Province
- BK20190185/Natural Science Foundation of Jiangsu Province
- BK20190186/Natural Science Foundation of Jiangsu Province
- BK20191175/Natural Science Foundation of Jiangsu Province
- No.16KJB310014/the Universities Natural Science Foundation of Jiangsu Province
- BE2021657/Jiangsu province's science and technology support program (Social Development)
- BE2021654/Jiangsu province's science and technology support program (Social Development) project
- BE2020659/Jiangsu Province Key R&D Program (Social Development) Projects
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

