In silico evaluation of a targeted metaproteomics strategy for broad screening of cellulolytic enzyme capacities in anaerobic microbiome bioreactors
- PMID: 35303956
- PMCID: PMC8933973
- DOI: 10.1186/s13068-022-02125-x
In silico evaluation of a targeted metaproteomics strategy for broad screening of cellulolytic enzyme capacities in anaerobic microbiome bioreactors
Abstract
Background: Microbial-driven solubilization of lignocellulosic material is a natural mechanism that is exploited in anaerobic digesters (ADs) to produce biogas and other valuable bioproducts. Glycoside hydrolases (GHs) are the main enzymes that bacterial and archaeal populations use to break down complex polysaccharides in these reactors. Methodologies for rapidly screening the physical presence and types of GHs can provide information about their functional activities as well as the taxonomical diversity within AD systems but are largely unavailable. Targeted proteomic methods could potentially be used to provide snapshots of the GHs expressed by microbial consortia in ADs, giving valuable insights into the functional lignocellulolytic degradation diversity of a community. Such observations would be essential to evaluate the hydrolytic performance of a reactor or potential issues with it.
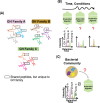
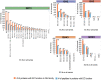
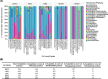
Results: As a proof of concept, we performed an in silico selection and evaluation of groups of tryptic peptides from five important GH families derived from a dataset of 1401 metagenome-assembled genomes (MAGs) in anaerobic digesters. Following empirical rules of peptide-based targeted proteomics, we selected groups of shared peptides among proteins within a GH family while at the same time being unique compared to all other background proteins. In particular, we were able to identify a tractable unique set of peptides that were sufficient to monitor the range of GH families. While a few thousand peptides would be needed for comprehensive characterization of the main GH families, we found that at least 50% of the proteins in these families (such as the key families) could be tracked with only 200 peptides. The unique peptides selected for groups of GHs were found to be sufficient for distinguishing enzyme specificity or microbial taxonomy. These in silico results demonstrate the presence of specific unique GH peptides even in a highly diverse and complex microbiome and reveal the potential for development of targeted metaproteomic approaches in ADs or lignocellulolytic microbiomes. Such an approach could be valuable for estimating molecular-level enzymatic capabilities and responses of microbial communities to different substrates or conditions, which is a critical need in either building or utilizing constructed communities or defined cultures for bio-production.
Conclusions: This in silico study demonstrates the peptide selection strategy for quantifying relevant groups of GH proteins in a complex anaerobic microbiome and encourages the development of targeted metaproteomic approaches in fermenters. The results revealed that targeted metaproteomics could be a feasible approach for the screening of cellulolytic enzyme capacities for a range of anaerobic microbiome fermenters and thus could assist in bioreactor evaluation and optimization.
Keywords: Anaerobic digester; Biogas; Glycoside hydrolases; Lignocellulose; Microbial community; Microbiome; Peptides; Targeted metaproteomics.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Fardin JF, de Barros Jr O, Dias AP. Advances in renewable energies and power technologies. Elsevier; 2018. Biomass: some basics and biogas; pp. 1–37.
-
- Kainthola J, Kalamdhad AS, Goud VV. A review on enhanced biogas production from anaerobic digestion of lignocellulosic biomass by different enhancement techniques. Process Biochem. 2019;84:81–90.
-
- Pramanik SK, Suja FB, Zain SM, Pramanik BKJBTR. The anaerobic digestion process of biogas production from food waste: prospects and constraints. Bioresour Technol Rep. 2019;8:100310.
-
- Rasapoor M, Young B, Brar R, Sarmah A, Zhuang WQ, Baroutian S. Recognizing the challenges of anaerobic digestion: critical steps toward improving biogas generation. Fuel. 2020 doi: 10.1016/j.fuel.2019.116497. - DOI
-
- Azman S, Khadem AF, Van Lier JB, Zeeman G, Plugge CM. Presence and role of anaerobic hydrolytic microbes in conversion of lignocellulosic biomass for biogas production. Crit Rev Env Sci Technol. 2015;45(23):2523–2564.
LinkOut - more resources
Full Text Sources
Research Materials
