Tracking SARS-CoV-2 variants by entire S-gene analysis using long-range RT-PCR and Sanger sequencing
- PMID: 35304093
- PMCID: PMC8923710
- DOI: 10.1016/j.cca.2022.03.014
Tracking SARS-CoV-2 variants by entire S-gene analysis using long-range RT-PCR and Sanger sequencing
Abstract
Introduction: Genomic surveillance of the SARS-CoV-2 virus is important to assess transmissibility, disease severity, and vaccine effectiveness. The SARS-CoV-2 genome consists of approximately 30 kb single-stranded RNA that is too large to analyze the whole genome by Sanger sequencing. Thus, in this study, we performed Sanger sequencing following long-range RT-PCR of the entire SARS-CoV-2 S-gene and analyzed the mutational dynamics.
Methods: The 4 kb region, including the S-gene, was amplified by two-step long-range RT-PCR. Then, the entire S-gene sequence was determined by Sanger sequencing. The amino acid mutations were identified as compared with the reference SARS-CoV-2 genome.
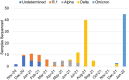
Results: The S:D614G mutation was found in all samples. The R.1 variants were detected after January 2021. Alpha variants started to emerge in April 2021. Delta variants replaced Alpha in July 2021. Then, Omicron variants were detected after December 2021. These mutational dynamics in samples collected in the Chiba University Hospital were similar to those in Japan.
Conclusion: The emergence of variants of concern (VOC) has been reported by the entire S-gene analysis. As the VOCs have unique mutational patterns of the S-gene region, analysis of the entire S-gene will be useful for molecular surveillance of the SARS-CoV-2 in clinical laboratories.
Keywords: Long-range RT-PCR; S-gene; SARS-CoV-2; Sanger sequencing; Surveillance.
Copyright © 2022 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
-
- World Health Organization (WHO), WHO Coronavirus (COVID-19) Dashboard. https://covid19.who.int, 2021 (accessed 16 December 2021).
-
- Volz E., Mishra S., Chand M., Barrett J.C., Johnson R., Geidelberg L., Hinsley W.R., Laydon D.J., Dabrera G., O’Toole Á., Amato R., Ragonnet-Cronin M., Harrison I., Jackson B., Ariani C.V., Boyd O., Loman N.J., McCrone J.T., Gonçalves S., Jorgensen D., Myers R., Hill V., Jackson D.K., Gaythorpe K., Groves N., Sillitoe J., Kwiatkowski D.P., Flaxman S., Ratmann O., Bhatt S., Hopkins S., Gandy A., Rambaut A., Ferguson N.M. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature. 2021;593(7858):266–269. - PubMed
-
- Singh J., Rahman S.A., Ehtesham N.Z., Hira S., Hasnain S.E. SARS-CoV-2 variants of concern are emerging in India. Nat. Med. 2021;27:1131–1133. - PubMed
-
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., Bailey R., Swanson K.A., Roychoudhury S., Koury K., Li P., Kalina W.V., Cooper D., Frenck R.W., Jr, Hammitt L.L., Türeci Ö., Nell H., Schaefer A., Ünal S., Tresnan D.B., Mather S., Dormitzer P.R., Şahin U., Jansen K.U., Gruber W.C. C4591001 Clinical Trial Group. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. - PMC - PubMed
-
- Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., McGettigan J., Khetan S., Segall N., Solis J., Brosz A., Fierro C., Schwartz H., Neuzil K., Corey L., Gilbert P., Janes H., Follmann D., Marovich M., Mascola J., Polakowski L., Ledgerwood J., Graham B.S., Bennett H., Pajon R., Knightly C., Leav B., Deng W., Zhou H., Han S., Ivarsson M., Miller J., Zaks T., COVE Study Group Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021;384:403–416. - PMC - PubMed
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

