Single-cell RNA sequencing of mast cells in eosinophilic esophagitis reveals heterogeneity, local proliferation, and activation that persists in remission
- PMID: 35304158
- PMCID: PMC9177790
- DOI: 10.1016/j.jaci.2022.02.025
Single-cell RNA sequencing of mast cells in eosinophilic esophagitis reveals heterogeneity, local proliferation, and activation that persists in remission
Abstract
Background: Mast cells (MCs) are pleiotropic cells that accumulate in the esophagus of patients with eosinophilic esophagitis (EoE) and are thought to contribute to disease pathogenesis, yet their properties and functions in this organ are largely unknown.
Objectives: This study aimed to perform a comprehensive molecular and spatial characterization of esophageal MCs in EoE.
Methods: Esophageal biopsies obtained from patients with active EoE, patients with EoE in histologic remission, and individuals with histologically normal esophageal biopsies and no history of esophageal disease (ie, control individuals) were subject to single-cell RNA sequencing, flow cytometry, and immunofluorescence analyses.
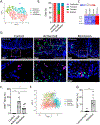
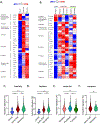
Results: This study probed 39,562 single esophageal cells by single-cell RNA sequencing; approximately 5% of these cells were MCs. Dynamic MC expansion was identified across disease states. During homeostasis, TPSAB1highAREGhigh resident MCs were mainly detected in the lamina propria and exhibited a quiescent phenotype. In patients with active EoE, resident MCs assumed an activated phenotype, and 2 additional proinflammatory MC populations emerged in the intraepithelial compartment, each linked to a proliferating MKI67high cluster. One proinflammatory activated MC population, marked as KIThighIL1RL1highFCER1Alow, was not detected in disease remission (termed "transient MC"), whereas the other population, marked as CMA1highCTSGhigh, was detected in disease remission where it maintained an activated state (termed "persistent MC"). MCs were prominent producers of esophageal IL-13 mRNA and protein, a key therapeutic target in EoE.
Conclusions: Esophageal MCs comprise heterogeneous populations with transcriptional signatures associated with distinct spatial compartmentalization and EoE disease status. In active EoE, they assume a proinflammatory state and locally proliferate, and they remain activated and poised to reinitiate inflammation even during disease remission.
Keywords: Eosinophilic esophagitis; IL-13; mast cells; proliferation; single-cell RNA sequencing.
Copyright © 2022 American Academy of Allergy, Asthma & Immunology. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Richter JE. Endoscopic Treatment of Eosinophilic Esophagitis. Gastrointest Endosc Clin N Am 2018; 28:97–110. - PubMed
-
- Schoepfer AM, Straumann A, Safroneeva E. Pharmacologic Treatment of Eosinophilic Esophagitis: An Update. Gastrointest Endosc Clin N Am 2018; 28:77–88. - PubMed
-
- Schoepfer AM, Gonsalves N, Bussmann C, Conus S, Simon HU, Straumann A, et al. Esophageal dilation in eosinophilic esophagitis: effectiveness, safety, and impact on the underlying inflammation. Am J Gastroenterol 2010; 105:1062–70. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

