Evolutionary transition from a single RNA replicator to a multiple replicator network
- PMID: 35304447
- PMCID: PMC8933500
- DOI: 10.1038/s41467-022-29113-x
Evolutionary transition from a single RNA replicator to a multiple replicator network
Abstract
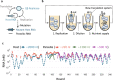
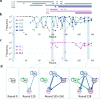
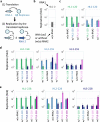
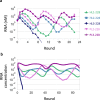
In prebiotic evolution, self-replicating molecules are believed to have evolved into complex living systems by expanding their information and functions open-endedly. Theoretically, such evolutionary complexification could occur through successive appearance of novel replicators that interact with one another to form replication networks. Here we perform long-term evolution experiments of RNA that replicates using a self-encoded RNA replicase. The RNA diversifies into multiple coexisting host and parasite lineages, whose frequencies in the population initially fluctuate and gradually stabilize. The final population, comprising five RNA lineages, forms a replicator network with diverse interactions, including cooperation to help the replication of all other members. These results support the capability of molecular replicators to spontaneously develop complexity through Darwinian evolution, a critical step for the emergence of life.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Comment in
-
In vitro evolution: From monsters to mobs.Curr Biol. 2022 Jun 20;32(12):R580-R583. doi: 10.1016/j.cub.2022.05.008. Curr Biol. 2022. PMID: 35728532
References
-
- Gilbert W. The RNA world. Nature. 1986;319:618.
-
- Szostak JW, Bartel DP, Luisi PL. Synthesizing life. Nature. 2001;409:387–390. - PubMed
-
- Joyce GF. The antiquity of RNA-based evolution. Nature. 2002;418:214–221. - PubMed
-
- Eigen M, Schuster P. A principle of natural self-organization—Part A: emergence of the hypercycle. Naturwissenschaften. 1977;64:541–565. - PubMed
-
- Maynard-Smith, J. & Szathmáry, E. The Major Transitions in Evolution (Oxford University Press, Oxford, UK, 1997).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

