S100A9-CXCL12 activation in BRCA1-mutant breast cancer promotes an immunosuppressive microenvironment associated with resistance to immunotherapy
- PMID: 35304461
- PMCID: PMC8933470
- DOI: 10.1038/s41467-022-29151-5
S100A9-CXCL12 activation in BRCA1-mutant breast cancer promotes an immunosuppressive microenvironment associated with resistance to immunotherapy
Abstract
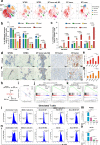
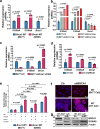
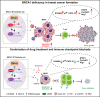
Immune checkpoint blockade (ICB) is a powerful approach for cancer therapy although good responses are only observed in a fraction of cancer patients. Breast cancers caused by deficiency of breast cancer-associated gene 1 (BRCA1) do not have an improved response to the treatment. To investigate this, here we analyze BRCA1 mutant mammary tissues and tumors derived from both BRCA1 mutant mouse models and human xenograft models to identify intrinsic determinants governing tumor progression and ICB responses. We show that BRCA1 deficiency activates S100A9-CXCL12 signaling for cancer progression and triggers the expansion and accumulation of myeloid-derived suppressor cells (MDSCs), creating a tumor-permissive microenvironment and rendering cancers insensitive to ICB. These oncogenic actions can be effectively suppressed by the combinatory treatment of inhibitors for S100A9-CXCL12 signaling with αPD-1 antibody. This study provides a selective strategy for effective immunotherapy in patients with elevated S100A9 and/or CXCL12 protein levels.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Narod SA, Foulkes WD. BRCA1 and BRCA2: 1994 and beyond. Nat. Rev. Cancer. 2004;4:665–676. - PubMed
-
- Takaoka M, Miki Y. BRCA1 gene: function and deficiency. Int J. Clin. Oncol. 2018;23:36–44. - PubMed
-
- Sharma B, Preet Kaur R, Raut S, Munshi A. BRCA1 mutation spectrum, functions, and therapeutic strategies: the story so far. Curr. Probl. Cancer. 2018;42:189–207. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

