The differential diagnosis of IgG4-related disease based on machine learning
- PMID: 35305690
- PMCID: PMC8933663
- DOI: 10.1186/s13075-022-02752-7
The differential diagnosis of IgG4-related disease based on machine learning
Abstract
Introduction: To eliminate the disparity and maldistribution of physicians and medical specialty services, the development of diagnostic support for rare diseases using artificial intelligence is being promoted. Immunoglobulin G4 (IgG4)-related disease (IgG4-RD) is a rare disorder often requiring special knowledge and experience to diagnose. In this study, we investigated the possibility of differential diagnosis of IgG4-RD based on basic patient characteristics and blood test findings using machine learning.
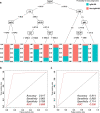
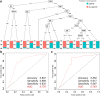
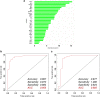
Methods: Six hundred and two patients with IgG4-RD and 204 patients with non-IgG4-RD that needed to be differentiated who visited the participating institutions were included in the study. Ten percent of the subjects were randomly excluded as a validation sample. Among the remaining cases, 80% were used as training samples, and the remaining 20% were used as test samples. Finally, validation was performed on the validation sample. The analysis was performed using a decision tree and a random forest model. Furthermore, a comparison was made between conditions with and without the serum IgG4 concentration. Accuracy was evaluated using the area under the receiver-operating characteristic (AUROC) curve.
Results: In diagnosing IgG4-RD, the AUROC curve values of the decision tree and the random forest method were 0.906 and 0.974, respectively, when serum IgG4 levels were included in the analysis. Excluding serum IgG4 levels, the AUROC curve value of the analysis by the random forest method was 0.925.
Conclusion: Based on machine learning in a multicenter collaboration, with or without serum IgG4 data, basic patient characteristics and blood test findings alone were sufficient to differentiate IgG4-RD from non-IgG4-RD.
Keywords: Artificial intelligence; Differential diagnosis; IgG4-related disease; Machine learning.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

