Coordination of SARS-CoV-2 wastewater and clinical testing of university students demonstrates the importance of sampling duration and collection time
- PMID: 35306079
- PMCID: PMC8925087
- DOI: 10.1016/j.scitotenv.2022.154619
Coordination of SARS-CoV-2 wastewater and clinical testing of university students demonstrates the importance of sampling duration and collection time
Abstract
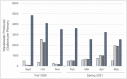
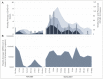
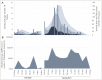
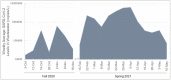
Wastewater surveillance has been a useful tool complementing clinical testing during the COVID-19 pandemic. However, transitioning surveillance approaches to small populations, such as dormitories and assisted living facilities poses challenges including difficulties with sample collection and processing. Recently, the need for reliable and timely data has coincided with the need for precise local forecasting of the trajectory of COVID-19. This study compared wastewater and clinical data from the University of Delaware (Fall 2020 and Spring 2021 semesters), and evaluated wastewater collection practices for enhanced virus detection sensitivity. Fecal shedding of SARS-CoV-2 is known to occur in infected individuals. However, shedding concentrations and duration has been shown to vary. Therefore, three shedding periods (14, 21, and 30 days) were presumed and included for analysis of wastewater data. SARS-CoV-2 levels detected in wastewater correlated with clinical virus detection when a positive clinical test result was preceded by fecal shedding of 21 days (p< 0.05) and 30 days (p < 0.05), but not with new cases (p = 0.09) or 14 days of shedding (p = 0.17). Discretely collected wastewater samples were compared with 24-hour composite samples collected at the same site. The discrete samples (n = 99) were composited examining the influence of sampling duration and time of day on SARS-CoV-2 detection. SARS-CoV-2 detection varied among dormitory complexes and sampling durations of 3-hour, 12-hour, and 24-hour (controls). Collection times frequently showing high detection values were between the hours of 03:00 to 05:00 and 23:00 to 08:00. In each of these times of day 33% of samples (3/9) were significantly higher (p < 0.05) than the control sample. The remainder (6/9) of the collection times (3-hour and 12-hour) were not different (p > 0.05) from the control. This study provides additional framework for continued methodology development for microbiological wastewater surveillance as the COVID-19 pandemic progresses and in preparation for future epidemiological efforts.
Keywords: COVID-19; Method development; SARS-CoV-2; Virus recovery; Wastewater-based epidemiology.
Copyright © 2022 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
-
- Abdeldayem O.M., Dabbish A.M., Habashy M.M., Mostafa M.K., Elhefnawy M., Amin L., Al-Sakkari E.G., Ragab A., Rene E.R. Viral outbreaks detection and surveillance using wastewater-based epidemiology, viral air sampling, and machine learning techniques: a comprehensive review and outlook. Sci. Total Environ. 2021;803 doi: 10.1016/j.scitotenv.2021.149834. - DOI - PMC - PubMed
-
- Betancourt W.Q., Schmitz B.W., Innes G.K., Prasek S.M., Brown K.M.P., Stark E.R., Foster A.R., Sprissler R.S., Harris D.T., Sherchan S.P., Gerba C.P., Pepper I.L. COVID-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention. Sci. Total Environ. 2021;779(146408):1–8. doi: 10.1016/j.scitotenv.2021.146408. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

