A role for the periplasmic adaptor protein AcrA in vetting substrate access to the RND efflux transporter AcrB
- PMID: 35306531
- PMCID: PMC8934357
- DOI: 10.1038/s41598-022-08903-9
A role for the periplasmic adaptor protein AcrA in vetting substrate access to the RND efflux transporter AcrB
Abstract
Tripartite resistance-nodulation-division (RND) efflux pumps, such as AcrAB-TolC of Salmonella Typhimurium, contribute to antibiotic resistance and comprise an inner membrane RND-transporter, an outer membrane factor, and a periplasmic adaptor protein (PAP). The role of the PAP in the assembly and active transport process remains poorly understood. Here, we identify the functionally critical residues involved in PAP-RND-transporter binding between AcrA and AcrB and show that the corresponding RND-binding residues in the closely related PAP AcrE, are also important for its interaction with AcrB. We also report a residue in the membrane-proximal domain of AcrA, that when mutated, differentially affects the transport of substrates utilising different AcrB efflux channels, namely channels 1 and 2. This supports a potential role for the PAP in sensing the substrate-occupied state of the proximal binding pocket of the transporter and substrate vetting. Understanding the PAP's role in the assembly and function of tripartite RND pumps can guide novel ways to inhibit their function to combat antibiotic resistance.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


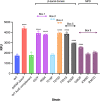

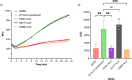
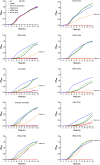
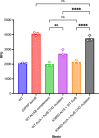
References
-
- Blair JM, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJ. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015;13:42–51. - PubMed
-
- Du D, et al. Multidrug efflux pumps: Structure, function and regulation. Nat. Rev. Microbiol. 2018;16:523–539. - PubMed
-
- Colclough AL, et al. RND efflux pumps in Gram-negative bacteria; regulation, structure and role in antibiotic resistance. Future Microbiol. 2020;15:143–157. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

