Machine Learning of All Mycobacterium tuberculosis H37Rv RNA-seq Data Reveals a Structured Interplay between Metabolism, Stress Response, and Infection
- PMID: 35306876
- PMCID: PMC9044949
- DOI: 10.1128/msphere.00033-22
Machine Learning of All Mycobacterium tuberculosis H37Rv RNA-seq Data Reveals a Structured Interplay between Metabolism, Stress Response, and Infection
Abstract
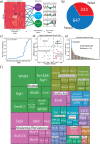
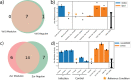
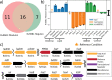
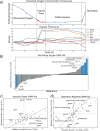
Mycobacterium tuberculosis is one of the most consequential human bacterial pathogens, posing a serious challenge to 21st century medicine. A key feature of its pathogenicity is its ability to adapt its transcriptional response to environmental stresses through its transcriptional regulatory network (TRN). While many studies have sought to characterize specific portions of the M. tuberculosis TRN, and some studies have performed system-level analysis, few have been able to provide a network-based model of the TRN that also provides the relative shifts in transcriptional regulator activity triggered by changing environments. Here, we compiled a compendium of nearly 650 publicly available, high quality M. tuberculosis RNA-sequencing data sets and applied an unsupervised machine learning method to obtain a quantitative, top-down TRN. It consists of 80 independently modulated gene sets known as "iModulons," 41 of which correspond to known regulons. These iModulons explain 61% of the variance in the organism's transcriptional response. We show that iModulons (i) reveal the function of poorly characterized regulons, (ii) describe the transcriptional shifts that occur during environmental changes such as shifting carbon sources, oxidative stress, and infection events, and (iii) identify intrinsic clusters of regulons that link several important metabolic systems, including lipid, cholesterol, and sulfur metabolism. This transcriptome-wide analysis of the M. tuberculosis TRN informs future research on effective ways to study and manipulate its transcriptional regulation and presents a knowledge-enhanced database of all published high-quality RNA-seq data for this organism to date. IMPORTANCE Mycobacterium tuberculosis H37Rv is one of the world's most impactful pathogens, and a large part of the success of the organism relies on the differential expression of its genes to adapt to its environment. The expression of the organism's genes is driven primarily by its transcriptional regulatory network, and most research on the TRN focuses on identifying and quantifying clusters of coregulated genes known as regulons. While previous studies have relied on molecular measurements, in the manuscript we utilized an alternative technique that performs machine learning to a large data set of transcriptomic data. This approach is less reliant on hypotheses about the role of specific regulatory systems and allows for the discovery of new biological findings for already collected data. A better understanding of the structure of the M. tuberculosis TRN will have important implications in the design of improved therapeutic approaches.
Keywords: Mycobacterium tuberculosis; gene regulation; independent component analysis; machine learning; transcriptomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- World Health Organization. GLOBAL TUBERCULOSIS REPORT 2020. 2020. Available: https://www.who.int/news-room/fact-sheets/detail/tuberculosis.
-
- Galagan JE, Minch K, Peterson M, Lyubetskaya A, Azizi E, Sweet L, Gomes A, Rustad T, Dolganov G, Glotova I, Abeel T, Mahwinney C, Kennedy AD, Allard R, Brabant W, Krueger A, Jaini S, Honda B, Yu W-H, Hickey MJ, Zucker J, Garay C, Weiner B, Sisk P, Stolte C, Winkler JK, Van de Peer Y, Iazzetti P, Camacho D, Dreyfuss J, Liu Y, Dorhoi A, Mollenkopf H-J, Drogaris P, Lamontagne J, Zhou Y, Piquenot J, Park ST, Raman S, Kaufmann SHE, Mohney RP, Chelsky D, Moody DB, Sherman DR, Schoolnik GK. 2013. The Mycobacterium tuberculosis regulatory network and hypoxia. Nature 499:178–183. doi:10.1038/nature12337. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

