Low-frequency variants in mildly symptomatic vaccine breakthrough infections presents a doubled-edged sword
- PMID: 35307848
- PMCID: PMC9325371
- DOI: 10.1002/jmv.27726
Low-frequency variants in mildly symptomatic vaccine breakthrough infections presents a doubled-edged sword
Abstract
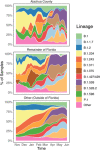
The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern (VOC) has raised questions regarding vaccine protection against SARS-CoV-2 infection, transmission, and ongoing virus evolution. Twenty-three mildly symptomatic "vaccination breakthrough" infections were identified as early as January 2021 in Alachua County, Florida, among individuals fully vaccinated with either the BNT162b2 (Pfizer) or the Ad26 (Janssen/J&J) vaccines. SARS-CoV-2 genomes were successfully generated for 11 of the vaccine breakthroughs, and 878 individuals in the surrounding area and were included for reference-based phylogenetic investigation. These 11 individuals were characterized by infection with VOCs, but also low-frequency variants present within the surrounding population. Low-frequency mutations were observed, which have been more recently identified as mutations of interest owing to their location within targeted immune epitopes (P812L) and association with increased replicative capacity (L18F). We present these results to posit the nature of the efficacy of vaccines in reducing symptoms as both a blessing and a curse-as vaccination becomes more widespread and self-motivated testing reduced owing to the absence of severe symptoms, we face the challenge of early recognition of novel mutations of potential concern. This case study highlights the critical need for continued testing and monitoring of infection and transmission among individuals regardless of vaccination status.
Keywords: SARS coronavirus; epidemiology; genetic variation; genetics; virus classification.
© 2022 The Authors. Journal of Medical Virology published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Comment in
-
The complex evolutionary dynamics of SARS-CoV-2, a big challenge to control the pandemic of COVID-19.J Med Virol. 2022 Nov;94(11):5082-5085. doi: 10.1002/jmv.27979. Epub 2022 Jul 11. J Med Virol. 2022. PMID: 35798562 Free PMC article. No abstract available.
References
-
- Rambaut A, Loman N, Pybus O, et al. Preliminary genomic characterisation of an emergent SARS‐CoV‐2 lineage in the UK defined by a novel set of spike mutations. Virologicalorg; 2020. https://virological.org/t/preliminary-genomic-characterisation-of-an-eme...
-
- CDC . SARS‐CoV‐2 Variant Classifications and Definitions. Accessed on February 9, 2022. https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classificatio...
-
- Larsen B, Worobey M. Phylogenetic evidence that B.1.1.7 has been circulating in the United States since early‐ to mid‐November. Virologicalorg. 2021. (https://virological.org/t/phylogenetic-evidence-that-b-1-1-7-has-been-ci...)
-
- Tegally H, Wilkinson E, Giovanetti M, et al. Detection of a SARS‐CoV‐2 variant of concern in South Africa. Nature. 2021;592(7854):438‐443. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

