Structure of the Pf12 and Pf41 heterodimeric complex of Plasmodium falciparum 6-cysteine proteins
- PMID: 35308105
- PMCID: PMC8930183
- DOI: 10.1093/femsmc/xtac005
Structure of the Pf12 and Pf41 heterodimeric complex of Plasmodium falciparum 6-cysteine proteins
Abstract
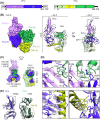
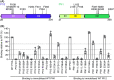
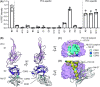
During the different stages of the Plasmodium life cycle, surface-associated proteins establish key interactions with the host and play critical roles in parasite survival. The 6-cysteine (6-cys) protein family is one of the most abundant surface antigens and expressed throughout the Plasmodium falciparum life cycle. This protein family is conserved across Plasmodium species and plays critical roles in parasite transmission, evasion of the host immune response and host cell invasion. Several 6-cys proteins are present on the parasite surface as hetero-complexes but it is not known how two 6-cys proteins interact together. Here, we present a crystal structure of Pf12 bound to Pf41 at 2.85 Å resolution, two P. falciparum proteins usually found on the parasite surface of late schizonts and merozoites. Our structure revealed two critical interfaces required for complex formation with important implications on how different 6-cysteine proteins may interact with each other. Using structure-function analyses, we identified important residues for Pf12-Pf41 complex formation. In addition, we generated 16 nanobodies against Pf12 and Pf41 and showed that several Pf12-specific nanobodies inhibit Pf12-Pf41 complex formation. Using X-ray crystallography, we were able to describe the structural mechanism of an inhibitory nanobody in blocking Pf12-Pf41 complex formation. Future studies using these inhibitory nanobodies will be useful to determine the functional role of these two 6-cys proteins in malaria parasites.
Keywords: 6-cysteine proteins; Nanobodies; Plasmodium falciparum; X-ray crystallography; blood stages; malaria.
© The Author(s) 2022. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
None declared.
Figures

Similar articles
-
Plasmodium 6-Cysteine Proteins: Functional Diversity, Transmission-Blocking Antibodies and Structural Scaffolds.Front Cell Infect Microbiol. 2022 Jul 8;12:945924. doi: 10.3389/fcimb.2022.945924. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35899047 Free PMC article. Review.
-
The Structure of Plasmodium falciparum Blood-Stage 6-Cys Protein Pf41 Reveals an Unexpected Intra-Domain Insertion Required for Pf12 Coordination.PLoS One. 2015 Sep 28;10(9):e0139407. doi: 10.1371/journal.pone.0139407. eCollection 2015. PLoS One. 2015. PMID: 26414347 Free PMC article.
-
Structural and biochemical characterization of Plasmodium falciparum 12 (Pf12) reveals a unique interdomain organization and the potential for an antiparallel arrangement with Pf41.J Biol Chem. 2013 May 3;288(18):12805-17. doi: 10.1074/jbc.M113.455667. Epub 2013 Mar 19. J Biol Chem. 2013. PMID: 23511632 Free PMC article.
-
Nanobody generation and structural characterization of Plasmodium falciparum 6-cysteine protein Pf12p.Biochem J. 2021 Feb 12;478(3):579-595. doi: 10.1042/BCJ20200415. Biochem J. 2021. PMID: 33480416 Free PMC article.
-
The s48/45 six-cysteine proteins: mediators of interaction throughout the Plasmodium life cycle.Int J Parasitol. 2017 Jun;47(7):409-423. doi: 10.1016/j.ijpara.2016.10.002. Epub 2016 Nov 27. Int J Parasitol. 2017. PMID: 27899328 Free PMC article. Review.
Cited by
-
Structure of endogenous Pfs230:Pfs48/45 in complex with potent malaria transmission-blocking antibodies.bioRxiv [Preprint]. 2025 Jun 15:2025.02.14.638310. doi: 10.1101/2025.02.14.638310. bioRxiv. 2025. PMID: 39990443 Free PMC article. Preprint.
-
Human coronavirus OC43 nanobody neutralizes virus and protects mice from infection.J Virol. 2024 Jun 13;98(6):e0053124. doi: 10.1128/jvi.00531-24. Epub 2024 May 6. J Virol. 2024. PMID: 38709106 Free PMC article.
-
PCRCR complex is essential for invasion of human erythrocytes by Plasmodium falciparum.Nat Microbiol. 2022 Dec;7(12):2039-2053. doi: 10.1038/s41564-022-01261-2. Epub 2022 Nov 17. Nat Microbiol. 2022. PMID: 36396942 Free PMC article.
-
Nanobodies against Pfs230 block Plasmodium falciparum transmission.Biochem J. 2022 Dec 22;479(24):2529-2546. doi: 10.1042/BCJ20220554. Biochem J. 2022. PMID: 36520108 Free PMC article.
-
Plasmodium 6-Cysteine Proteins: Functional Diversity, Transmission-Blocking Antibodies and Structural Scaffolds.Front Cell Infect Microbiol. 2022 Jul 8;12:945924. doi: 10.3389/fcimb.2022.945924. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35899047 Free PMC article. Review.
References
-
- Adams PD, Grosse-Kunstleve RW, Hung LWet al. . PHENIX: building new software for automated crystallographic structure determination. Acta Crystallographica Sect D Biolog Crystall. 2002;58:1948–54. - PubMed
-
- Annoura T, van Schaijk BC, Ploemen IHet al. . Two plasmodium 6-Cys family-related proteins have distinct and critical roles in liver-stage development. FASEB J. 2014;28:2158–70. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
