Force-Generation by the Trans-Envelope Tol-Pal System
- PMID: 35308353
- PMCID: PMC8928145
- DOI: 10.3389/fmicb.2022.852176
Force-Generation by the Trans-Envelope Tol-Pal System
Abstract
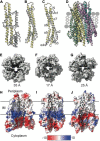
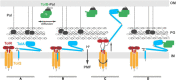
The Tol-Pal system spans the cell envelope of Gram-negative bacteria, transducing the potential energy of the proton motive force (PMF) into dissociation of the TolB-Pal complex at the outer membrane (OM), freeing the lipoprotein Pal to bind the cell wall. The primary physiological role of Tol-Pal is to maintain OM integrity during cell division through accumulation of Pal molecules at division septa. How the protein complex couples the PMF at the inner membrane into work at the OM is unknown. The effectiveness of this trans-envelope energy transduction system is underscored by the fact that bacteriocins and bacteriophages co-opt Tol-Pal as part of their import/infection mechanisms. Mechanistic understanding of this process has been hindered by a lack of structural data for the inner membrane TolQ-TolR stator, of its complexes with peptidoglycan (PG) and TolA, and of how these elements combined power events at the OM. Recent studies on the homologous stators of Ton and Mot provide a starting point for understanding how Tol-Pal works. Here, we combine ab initio protein modeling with previous structural data on sub-complexes of Tol-Pal as well as mutagenesis, crosslinking, co-conservation analysis and functional data. Through this composite pooling of in silico, in vitro, and in vivo data, we propose a mechanism for force generation in which PMF-driven rotary motion within the stator drives conformational transitions within a long TolA helical hairpin domain, enabling it to reach the TolB-Pal complex at the OM.
Keywords: Gram-negative bacteria; Tol-Pal; cell envelope; force transduction; outer membrane; proton motive force.
Copyright © 2022 Webby, Williams-Jones, Press and Kleanthous.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

