Comparing Long-Read Assemblers to Explore the Potential of a Sustainable Low-Cost, Low-Infrastructure Approach to Sequence Antimicrobial Resistant Bacteria With Oxford Nanopore Sequencing
- PMID: 35308384
- PMCID: PMC8928191
- DOI: 10.3389/fmicb.2022.796465
Comparing Long-Read Assemblers to Explore the Potential of a Sustainable Low-Cost, Low-Infrastructure Approach to Sequence Antimicrobial Resistant Bacteria With Oxford Nanopore Sequencing
Abstract
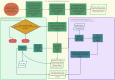
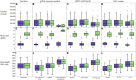
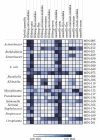
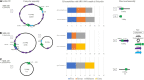
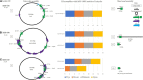
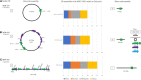
Long-read sequencing (LRS) can resolve repetitive regions, a limitation of short read (SR) data. Reduced cost and instrument size has led to a steady increase in LRS across diagnostics and research. Here, we re-basecalled FAST5 data sequenced between 2018 and 2021 and analyzed the data in relation to gDNA across a large dataset (n = 200) spanning a wide GC content (25-67%). We examined whether re-basecalled data would improve the hybrid assembly, and, for a smaller cohort, compared long read (LR) assemblies in the context of antimicrobial resistance (AMR) genes and mobile genetic elements. We included a cost analysis when comparing SR and LR instruments. We compared the R9 and R10 chemistries and reported not only a larger yield but increased read quality with R9 flow cells. There were often discrepancies with ARG presence/absence and/or variant detection in LR assemblies. Flye-based assemblies were generally efficient at detecting the presence of ARG on both the chromosome and plasmids. Raven performed more quickly but inconsistently recovered small plasmids, notably a ∼15-kb Col-like plasmid harboring bla KPC . Canu assemblies were the most fragmented, with genome sizes larger than expected. LR assemblies failed to consistently determine multiple copies of the same ARG as identified by the Unicycler reference. Even with improvements to ONT chemistry and basecalling, long-read assemblies can lead to misinterpretation of data. If LR data are currently being relied upon, it is necessary to perform multiple assemblies, although this is resource (computing) intensive and not yet readily available/useable.
Keywords: Guppy; MinION; Oxford Nanopore Technology (ONT); antimicrobial resistance (AMR); antimicrobial resistance genes (ARG); de novo assembly; long-read sequencing (LRS); plasmid.
Copyright © 2022 Boostrom, Portal, Spiller, Walsh and Sands.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Benton M. (2021). Nvidia Jetson Nanopore Sequencing: A Place to Collate Notes and Resources of Our Journey Into Porting Nanopore Sequencing Over to Accessible, Portable Technology. San Francisco, CA: GitHub.
-
- Cardiff University (2021). Cardiff University eMarketplace. Cardiff: Cardiff University.
-
- Cerdeira L. T., Lam M. M. C., Wyres K. L., Wick R. R., Judd L. M., Lopes R., et al. (2019). Small IncQ1 and col-like plasmids harboring blaKPC-2 and non-Tn4401 elements (NTEKPC-IId) in high-risk lineages of Klebsiella pneumoniae CG258. Antimicrob Agents Chemother. 63:e02140-18. 10.1128/AAC.02140-2118 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

