PD-1 N58-Glycosylation-Dependent Binding of Monoclonal Antibody Cemiplimab for Immune Checkpoint Therapy
- PMID: 35309324
- PMCID: PMC8924070
- DOI: 10.3389/fimmu.2022.826045
PD-1 N58-Glycosylation-Dependent Binding of Monoclonal Antibody Cemiplimab for Immune Checkpoint Therapy
Abstract
Immune checkpoint therapy (ICT) with a monoclonal antibody (MAb) against programmed cell death protein 1 (PD-1) is a powerful clinical treatment for tumors. Cemiplimab is a human IgG4 antibody approved in 2018 and is the first MAb proven to be effective for locally advanced basal cell carcinoma. Here, we report the crystal structure of cemiplimab bound to PD-1 and the effects of PD-1 N-glycosylation on the interactions with cemiplimab. The structure of the cemiplimab/PD-1 complex shows that cemiplimab mainly binds to PD-1 with its heavy chain, whereas the light chain serves as the predominant region to compete with the binding of PD-L1 to PD-1. The interaction network of cemiplimab to PD-1 resembles that of camrelizumab (another PD-1-binding MAb), and the N58 glycan on the BC loop of PD-1 may be involved in the interaction with cemiplimab. The binding affinity of cemiplimab with PD-1 was substantially decreased with N58-glycan-deficient PD-1, whereas the PD-1/PD-L1 blocking efficiency of cemiplimab was attenuated upon binding to the N58-glycosylation-deficient PD-1. These results indicate that both the binding and blocking efficacy of cemiplimab require the N58 glycosylation of PD-1. Taken together, these findings expand our understanding of the significance of PD-1 glycosylation in the interaction with cemiplimab.
Keywords: N58 glycosylation; PD-1; antibody; cemiplimab; immune checkpoint therapy (ICT).
Copyright © 2022 Lu, Xu, Zhang, Jiang, Liu, He, Ma, Ma, Tan, Gao and Chai.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


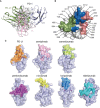

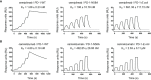

References
-
- Chae YK, Arya A, Iams W, Cruz MR, Chandra S, Choi J, et al. Current Landscape and Future of Dual Anti-CTLA4 and PD-1/PD-L1 Blockade Immunotherapy in Cancer; Lessons Learned From Clinical Trials With Melanoma and Non-Small Cell Lung Cancer (NSCLC). J Immunother Cancer (2018) 6(1):39. doi: 10.1186/s40425-018-0349-3 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

