Theoretical Development of DnaG Primase as a Novel Narrow-Spectrum Antibiotic Target
- PMID: 35309427
- PMCID: PMC8928506
- DOI: 10.1021/acsomega.1c05928
Theoretical Development of DnaG Primase as a Novel Narrow-Spectrum Antibiotic Target
Abstract
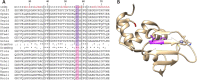
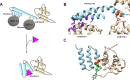
The widespread use of antibiotics to treat infections is one of the reasons that global mortality rates have fallen over the past 80 years. However, antibiotic use is also responsible for the concomitant rise in antibiotic resistance because it results in dysbiosis in which commensal and pathogenic bacteria are both greatly reduced. Therefore, narrow-range antibiotics are a promising direction for reducing antibiotic resistance because they are more discriminate. As a step toward addressing this problem, the goal of this study was to identify sites on DnaG primase that are conserved within Gram-positive bacteria and different from the equivalent sites in Gram-negative bacteria. Based on sequence and structural analysis, the primase C-terminal helicase-binding domain (CTD) was identified as most promising. Although the primase CTD sequences are very poorly conserved, they have highly conserved protein folds, and Gram-positive bacterial primases fold into a compact state that creates a small molecule binding site adjacent to a groove. The small molecule would stabilize the protein in its compact state, which would interfere with the helicase binding. This is important because primase CTD must be in its open conformation to bind to its cognate helicase at the replication fork.
© 2022 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures






References
-
- O’Neill J.Tackling Drug-resistant Infections Globally: Final Report and Recommendations. Review on Antimicrobial Resistance https://amr-review.org/sites/default/files/160525_Final%20paper_with%20c....
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

