Omicron's binding to sotrovimab, casirivimab, imdevimab, CR3022, and sera from previously infected or vaccinated individuals
- PMID: 35309727
- PMCID: PMC8920075
- DOI: 10.1016/j.isci.2022.104076
Omicron's binding to sotrovimab, casirivimab, imdevimab, CR3022, and sera from previously infected or vaccinated individuals
Abstract
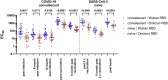
SARS-CoV-2 Omicron is the first pandemic variant of concern exhibiting an abrupt accumulation of mutations particularly in the receptor-binding domain that is a critical target of vaccination induced and therapeutic antibodies. Omicron's mutations did only marginally affect the binding of ACE2, and the two antibodies Sotrovimab and CR3022 but strongly impaired the binding of Casirivimab and Imdevimab. Moreover, as compared with Wuhan, there is reduced serum reactivity and a pronounced loss of competitive surrogate virus neutralization (sVN) against Omicron in naïve vaccinees and in COVID-19 convalescents after infection and subsequent vaccination. Finally, although the booster vaccination response conferred higher titers and better sVN, the effect was nonetheless significantly lower compared with responses against Wuhan. Overall, our data suggest that the antigenicity of Omicrons receptor binding motive has largely changed but antibodies such as Sotrovimab targeting other conserved sites maintain binding and therefore hold potential in prophylaxis and treatment of Omicron-induced COVID-19.
Keywords: Biological sciences; Health sciences; Immunology; Medicine.
© 2022 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Ai J., Zhang H., Zhang Y., Lin K., Zhang Y., Wu J., Wan Y., Huang Y., Song J., Fu Z., et al. Omicron variant showed lower neutralizing sensitivity than other SARS-CoV-2 variants to immune sera elicited by vaccines after boost. Emerg. Microbes Infect. 2021;11:337–343. doi: 10.1080/22221751.2021.2022440. - DOI - PMC - PubMed
-
- Andrews N., Stowe J., Kirsebom F., Toffa S., Rickeard T., Gallagher E., Gower C., Kall M., Groves N., O’Connell A.-M., et al. Effectiveness of COVID-19 vaccines against the Omicron (B.1.1.529) variant of concern. medRxiv. 2021 doi: 10.1101/2021.12.14.21267615. Preprint at. - DOI
-
- Asthagiri Arunkumar G., Ioannou A., Wohlbold T.J., Meade P., Aslam S., Amanat F., Ayllon J., García-Sastre A., Krammer F. Broadly cross-reactive, nonneutralizing antibodies against influenza B virus hemagglutinin demonstrate effector function-dependent protection against lethal viral challenge in mice. J. Virol. 2019;93:e01696-18. doi: 10.1128/JVI.01696-18. - DOI - PMC - PubMed
-
- Barnes C.O., Jette C.A., Abernathy M.E., Dam K.-M.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. Structural classification of neutralizing antibodies against the SARS-CoV-2 spike receptor-binding domain suggests vaccine and therapeutic strategies. bioRxiv. 2020 doi: 10.1101/2020.08.30.273920. Preprint at. - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

