Identification of the Ferroptosis-Related Long Non-Coding RNAs Signature to Improve the Prognosis Prediction in Papillary Renal Cell Carcinoma
- PMID: 35310430
- PMCID: PMC8930926
- DOI: 10.3389/fsurg.2022.741726
Identification of the Ferroptosis-Related Long Non-Coding RNAs Signature to Improve the Prognosis Prediction in Papillary Renal Cell Carcinoma
Abstract
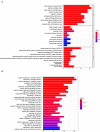
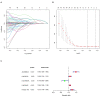
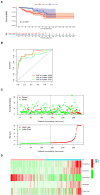
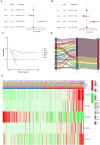
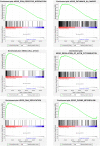
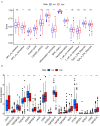
Papillary renal cell carcinoma (pRCC) is one of the epithelial renal cell carcinoma (RCC) histological subtypes. Ferroptosis is a new iron-dependent form of cell death that has been seen in a variety of clinical situations. Using differentially expressed ferroptosis-related long non-coding RNAs (lncRNAs) from patients with pRCC in The Cancer Genome Atlas; we built a prognostic lncRNA-based signature. We discovered seven different lncRNAs that were strongly linked to the prognosis of patients with pRCC. High-risk scores were linked to a poor prognosis for pRCC, which was confirmed by the findings of Kaplan-Meier studies. In addition, the constructed lncRNA signature has a 1-year area under the curve (AUC) of 0.908, suggesting that it has a high predictive value in pRCC. In the high-risk group, Gene set enrichment analyses (GSEA) analysis identified immunological and tumor-related pathways. Furthermore, single-sample GSEA (ssGSEA) revealed significant differences in T cell functions checkpoint, antigen presenting cell (APC) co-stimulation, inflammation promoting, and para inflammation between the two groups with different risk scores. In addition, immune checkpoints like PDCD1LG2 (PD-L2), LAG3, and IDO1 were expressed differently in the two risk groups. In summary, a novel signature based on ferroptosis-related lncRNAs could be applied in predicting the prognosis of patients with pRCC.
Keywords: ferroptosis; immune environment; lncRNA; papillary renal cell carcinoma; prognosis.
Copyright © 2022 Tang, Jiang, Wang, Xia, Mao and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer JL declared a shared affiliation, though no other collaboration, with the authors to the handling Editor.
Figures


References
LinkOut - more resources
Full Text Sources
Research Materials

