Ethylene Response Factor109 Attunes Immunity, Photosynthesis, and Iron Homeostasis in Arabidopsis Leaves
- PMID: 35310669
- PMCID: PMC8924546
- DOI: 10.3389/fpls.2022.841366
Ethylene Response Factor109 Attunes Immunity, Photosynthesis, and Iron Homeostasis in Arabidopsis Leaves
Abstract
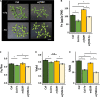
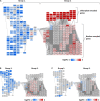
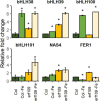
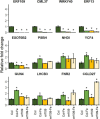
Iron (Fe) is an essential micronutrient element for all organisms including plants. Chlorosis of young leaves is a common symptom of Fe deficiency, reducing the efficiency of photosynthesis, and, ultimately, crop yield. Previous research revealed strong responsiveness of the putative key transcription factor ERF109 to the Fe regime. To elucidate the possible role of ERF109 in leaf Fe homeostasis and photosynthesis, we subjected Arabidopsis thaliana erf109 knockout lines and Col-0 wild-type plants to transcriptome profiling via RNA-seq. The transcriptome profile of Fe-sufficient erf109 leaves showed a 71% overlap with Fe-deficient Col-0 plants. On the other hand, genes that were differentially expressed between Fe-deficient and Fe-sufficient Col-0 plants remained unchanged in erf109 plants under conditions of Fe deficiency. Mutations in ERF109 increased the expression of the clade Ib bHLH proteins bHLH38, bHLH39, bHLH101, the nicotianamine synthase NAS4, and the Fe storage gene FER1. Moreover, mutations in ERF109 led to significant down-regulation of defense genes, including CML37, WRKY40, ERF13, and EXO70B2. Leaves of erf109 exhibited increased Fe levels under both Fe-sufficient and Fe-deficient conditions. Reduced Fv/Fm and Soil Plant Analysis Development (SPAD) values in erf109 lines under Fe deficiency indicate curtailed ability of photosynthesis relative to the wild-type. Our findings suggest that ERF109 is a negative regulator of the leaf response to Fe deficiency. It further appears that the function of ERF109 in the Fe response is critical for regulating pathogen defense and photosynthetic efficiency. Taken together, our study reveals a novel function of ERF109 and provides a systematic perspective on the intertwining of the immunity regulatory network and cellular Fe homeostasis.
Keywords: ERF109; RRTF1; immunity; iron deficiency; photosynthesis; transcriptome.
Copyright © 2022 Yang, Huang, Schmidt, Klein, Chan and Pan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Genome-wide and comparative analysis of bHLH38, bHLH39, bHLH100 and bHLH101 genes in Arabidopsis, tomato, rice, soybean and maize: insights into iron (Fe) homeostasis.Biometals. 2018 Aug;31(4):489-504. doi: 10.1007/s10534-018-0095-5. Epub 2018 Mar 15. Biometals. 2018. PMID: 29546482
-
Transcriptional coordination between leaf cell differentiation and chloroplast development established by TCP20 and the subgroup Ib bHLH transcription factors.Plant Mol Biol. 2014 Jun;85(3):233-45. doi: 10.1007/s11103-014-0180-2. Epub 2014 Feb 19. Plant Mol Biol. 2014. PMID: 24549883
-
FIT and bHLH Ib transcription factors modulate iron and copper crosstalk in Arabidopsis.Plant Cell Environ. 2021 May;44(5):1679-1691. doi: 10.1111/pce.14000. Epub 2021 Feb 3. Plant Cell Environ. 2021. PMID: 33464620
-
The Understanding of the Plant Iron Deficiency Responses in Strategy I Plants and the Role of Ethylene in This Process by Omic Approaches.Front Plant Sci. 2017 Jan 24;8:40. doi: 10.3389/fpls.2017.00040. eCollection 2017. Front Plant Sci. 2017. PMID: 28174585 Free PMC article. Review.
-
Regulation of the iron-deficiency response by IMA/FEP peptide.Front Plant Sci. 2023 Apr 25;14:1107405. doi: 10.3389/fpls.2023.1107405. eCollection 2023. Front Plant Sci. 2023. PMID: 37180394 Free PMC article. Review.
Cited by
-
Genome-wide analysis of AP2/ERF gene family and functional characterization of StoERF109 in Solanum torvum response to Verticillium dahliae infection.Planta. 2025 May 20;262(1):1. doi: 10.1007/s00425-025-04723-z. Planta. 2025. PMID: 40392396
-
Modulation of early gene expression responses to water deprivation stress by the E3 ubiquitin ligase ATL80: implications for retrograde signaling interplay.BMC Plant Biol. 2024 Mar 8;24(1):180. doi: 10.1186/s12870-024-04872-5. BMC Plant Biol. 2024. PMID: 38459432 Free PMC article.
-
Genome-wide association study of Verticillium longisporum resistance in Brassica genotypes.Front Plant Sci. 2024 Aug 27;15:1436982. doi: 10.3389/fpls.2024.1436982. eCollection 2024. Front Plant Sci. 2024. PMID: 39258297 Free PMC article.
-
Regulation of hormone pathways in wheat infested by Blumeria graminis f. sp. tritici.BMC Plant Biol. 2023 Nov 9;23(1):554. doi: 10.1186/s12870-023-04569-1. BMC Plant Biol. 2023. PMID: 37940874 Free PMC article.
-
GhERF.B4-15D: A Member of ERF Subfamily B4 Group Positively Regulates the Resistance against Verticillium dahliae in Upland Cotton.Biomolecules. 2023 Sep 5;13(9):1348. doi: 10.3390/biom13091348. Biomolecules. 2023. PMID: 37759747 Free PMC article.
References
-
- Àlvarez-Fernàndez A., Abadía J., Abadía A. (2006). “Iron deficiency, fruit yield and fruit quality,” in Iron Nutrition in Plants and Rhizospheric Microorganisms, eds Barton L. L., Abadia J. (Dordrecht: Springer Press; ), 85–101.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

