Metagenomic Assessment of the Pathogenic Risk of Microorganisms in Sputum of Postoperative Patients With Pulmonary Infection
- PMID: 35310849
- PMCID: PMC8928749
- DOI: 10.3389/fcimb.2022.855839
Metagenomic Assessment of the Pathogenic Risk of Microorganisms in Sputum of Postoperative Patients With Pulmonary Infection
Abstract
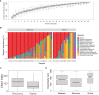
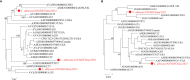
Respiratory infections are complicated biological processes associated with an unbalanced microbial community and a wide range of pathogens. To date, robust approaches are still required for distinguishing the pathogenic microorganisms from the colonizing ones in the clinical specimens with complex infection. In this study, we retrospectively analyzed the data of conventional culture testing and metagenomic next-generation sequencing (mNGS) of the sputum samples collected from 50 pulmonary infected patients after cardiac surgery from December 2020 and June 2021 in Ruijin Hospital. Taxonomic classification of the sputum metagenomes showed that the numbers of species belonging to bacteria, fungi, and viruses were 682, 58, and 21, respectively. The full spectrum of microorganisms present in the sputum microbiome covered all the species identified by culture, including 12 bacterial species and two fungal species. Based on species-level microbiome profiling, a reference catalog of microbial abundance detection limits was constructed to assess the pathogenic risks of individual microorganisms in the specimens. The proposed screening procedure detected 64 bacterial pathogens, 10 fungal pathogens, and three viruses. In particular, certain opportunistic pathogenic strains can be distinguished from the colonizing ones in the individual specimens. Strain-level identification and phylogenetic analysis were further performed to decipher molecular epidemiological characteristics of four opportunistic etiologic agents, including Klebsiella pneumoniae, Corynebacterium striatum, Staphylococcus aureus, and Candida albicans. Our findings provide a novel metagenomic insight into precision diagnosis for clinically relevant microbes, especially for opportunistic pathogens in the clinical setting.
Keywords: limit of detection; metagenomic next-generation sequencing; opportunistic pathogen; pathogen risk; pulmonary infection; sputum microbiome; strain profiling.
Copyright © 2022 Chen, Sun, Liu, Yu, Qin, Cao and Gu.
Conflict of interest statement
The authors LS and XC are employed by Genoxor Medical Science and Technology Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Adams H. M., Joyce L. R., Guan Z., Akins R. L., Palmer K. L. (2017). Streptococcus Mitis and S. Oralis Lack a Requirement for CdsA, the Enzyme Required for Synthesis of Major Membrane Phospholipids in Bacteria. Antimicrobial. Agents Chemother. 61 (5), e02552–e02516. doi: 10.1128/AAC.02552-16 - DOI - PMC - PubMed
-
- Alibi S., Ferjani A., Boukadida J., Cano M. E., Fernández-Martínez M., Martínez-Martínez L., et al. . (2017). Occurrence of Corynebacterium Striatum as an Emerging Antibiotic-Resistant Nosocomial Pathogen in a Tunisian Hospital. Sci. Rep. 7 (1), 9704. doi: 10.1038/s41598-017-10081-y - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

