The Involvement of Long Non-coding RNA and Messenger RNA Based Molecular Networks and Pathways in the Subacute Phase of Traumatic Brain Injury in Adult Mice
- PMID: 35311004
- PMCID: PMC8931714
- DOI: 10.3389/fninf.2022.794342
The Involvement of Long Non-coding RNA and Messenger RNA Based Molecular Networks and Pathways in the Subacute Phase of Traumatic Brain Injury in Adult Mice
Abstract
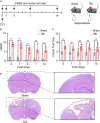
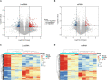
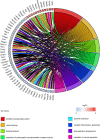
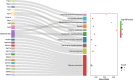
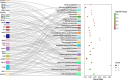
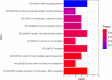
Traumatic brain injury (TBI) is a complex injury with a multi-faceted recovery process. Long non-coding RNAs (lncRNAs) are demonstrated to be involved in central nervous system (CNS) disorders. However, the roles of lncRNAs in long-term neurological deficits post-TBI are poorly understood. The present study depicted the microarray's lncRNA and messenger RNA (mRNA) profiles at 14 days in TBI mice hippocampi. LncRNA and mRNA microarray was used to identify differentially expressed genes. Quantitative real-time polymerase chain reaction (qRT-PCR) was employed to validate the microarray results. Bioinformatics analysis [including Gene Ontology (GO), the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, lncRNA-mRNA co-expression network, and lncRNA-miRNA-mRNA network] were applied to explore the underlying mechanism. A total of 264 differentially expressed lncRNAs and 232 expressed mRNAs were identified (fold change > 1.5 and P-value < 0.05). Altered genes were enriched in inflammation, immune response, blood-brain barrier, glutamatergic neurological effects, and neuroactive ligand-receptor, which may be associated with TBI-induced pathophysiologic changes in the long-term neurological deficits. The lncRNAs-mRNAs co-expression network was generated for 74 lncRNA-mRNA pairs, most of which are positive correlations. The lncRNA-miRNA-mRNA interaction network included 12 lncRNAs, 59 miRNAs, and 25 mRNAs. Numerous significantly altered lncRNAs and mRNAs in mice hippocampi were enriched in inflammation and immune response. Furthermore, these dysregulated lncRNAs and mRNAs may be promising therapeutic targets to overcome obstacles in long-term recovery following TBI.
Keywords: immune response; inflammation; lncRNA and mRNA microarray; subacute phase; traumatic brain injury.
Copyright © 2022 Yang, Li, Luo, Wu, Tang and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Altered Long Noncoding RNA and Messenger RNA Expression in Experimental Intracerebral Hemorrhage - a Preliminary Study.Cell Physiol Biochem. 2018;45(3):1284-1301. doi: 10.1159/000487464. Epub 2018 Feb 9. Cell Physiol Biochem. 2018. PMID: 29448258
-
Comprehensive Analysis of lncRNA Expression Pattern and lncRNA-miRNA-mRNA Network in a Rat Model With Cavernous Nerve Injury Erectile Dysfunction.J Sex Med. 2020 Sep;17(9):1603-1617. doi: 10.1016/j.jsxm.2020.05.008. Epub 2020 Jul 13. J Sex Med. 2020. PMID: 32675050
-
Characterization of the lncRNA-miRNA-mRNA regulatory network to reveal potential functional competing endogenous RNAs in traumatic brain injury.Front Neurosci. 2023 Jan 11;16:1089857. doi: 10.3389/fnins.2022.1089857. eCollection 2022. Front Neurosci. 2023. PMID: 36711143 Free PMC article.
-
The role of long noncoding RNA in traumatic brain injury.Neuropsychiatr Dis Treat. 2019 Jun 28;15:1671-1677. doi: 10.2147/NDT.S206624. eCollection 2019. Neuropsychiatr Dis Treat. 2019. PMID: 31303755 Free PMC article. Review.
-
Research Progress on the Inflammatory Effects of Long Non-coding RNA in Traumatic Brain Injury.Front Mol Neurosci. 2022 Mar 10;15:835012. doi: 10.3389/fnmol.2022.835012. eCollection 2022. Front Mol Neurosci. 2022. PMID: 35359568 Free PMC article. Review.
Cited by
-
Role of regulatory non-coding RNAs in traumatic brain injury.Neurochem Int. 2024 Jan;172:105643. doi: 10.1016/j.neuint.2023.105643. Epub 2023 Nov 24. Neurochem Int. 2024. PMID: 38007071 Free PMC article. Review.
-
Potential Correlation Between Molecular Biomarkers and Oxidative Stress in Traumatic Brain Injury.Int J Mol Sci. 2025 Apr 18;26(8):3858. doi: 10.3390/ijms26083858. Int J Mol Sci. 2025. PMID: 40332547 Free PMC article. Review.
-
YTHDF1 Attenuates TBI-Induced Brain-Gut Axis Dysfunction in Mice.Int J Mol Sci. 2023 Feb 20;24(4):4240. doi: 10.3390/ijms24044240. Int J Mol Sci. 2023. PMID: 36835655 Free PMC article.
References
LinkOut - more resources
Full Text Sources

