Immune Infiltration and Clinical Outcome of Super-Enhancer-Associated lncRNAs in Stomach Adenocarcinoma
- PMID: 35311149
- PMCID: PMC8927879
- DOI: 10.3389/fonc.2022.780493
Immune Infiltration and Clinical Outcome of Super-Enhancer-Associated lncRNAs in Stomach Adenocarcinoma
Abstract
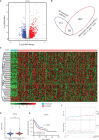
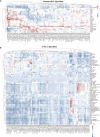
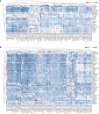
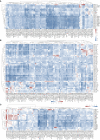
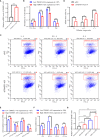
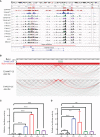
Super-enhancers (SEs) comprise large clusters of enhancers that highly enhance gene expression. Long non-coding RNAs (lncRNAs) tend to be dysregulated in cases of stomach adenocarcinoma (STAD) and are vital for balancing tumor immunity. However, whether SE-associated lncRNAs play a role in the immune infiltration of STAD remains unknown. In the present study, we identified SE-associated lncRNAs in the H3K27ac ChIP-seq datasets from 11 tumor tissues and two cell lines. We found that the significantly dysregulated SE-associated lncRNAs were strongly correlated with immune cell infiltration through the application of six algorithms (ImmuncellAI, CIBERSORT, EPIC, quantiSeq, TIMER, and xCELL), as well as immunomodulators and chemokines. We found that the expression of SE-associated lncRNA TM4SF1-AS1 was negatively correlated with the proportion of CD8+ T cells present in STAD. TM4SF1-AS1 suppresses T cell-mediated immune killing function and predicts immune response to anti-PD1 therapy. ChIP-seq, Hi-C and luciferase assay results verified that TM4SF1-AS1 was regulated by its super-enhancer. RNA-seq data showed that TM4SF1-AS1 is involved in immune and cancer-related processes or pathways. In conclusion, SE-associated lncRNAs are involved in the tumor immune microenvironment and act as indicators of clinical outcomes in STAD. This study highlights the importance of SE-associated lncRNAs in the immune regulation of STAD.
Keywords: PD1; T cell; TM4SF1-AS1; clinical outcome; immune infiltration; long non-coding RNA (IncRNA); stomach adenocarcinoma (STAD); super-enhancer (SE).
Copyright © 2022 Peng, Peng, Cai, Qiu, Lan, Luo, Yang, Xie, Du, Yuan, Liu and Yin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Nagaraja AK, Kikuchi O, Bass AJ. Genomics and Targeted Therapies in Gastroesophageal Adenocarcinoma. Cancer Discovery (2019) 9:1656–72. doi: 10.1158/2159-8290.cd-19-0487 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

