The GA4GH Variation Representation Specification: A computational framework for variation representation and federated identification
- PMID: 35311178
- PMCID: PMC8929418
- DOI: 10.1016/j.xgen.2021.100027
The GA4GH Variation Representation Specification: A computational framework for variation representation and federated identification
Abstract
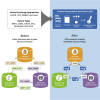
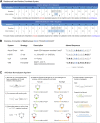
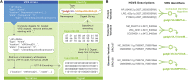
Maximizing the personal, public, research, and clinical value of genomic information will require the reliable exchange of genetic variation data. We report here the Variation Representation Specification (VRS, pronounced "verse"), an extensible framework for the computable representation of variation that complements contemporary human-readable and flat file standards for genomic variation representation. VRS provides semantically precise representations of variation and leverages this design to enable federated identification of biomolecular variation with globally consistent and unique computed identifiers. The VRS framework includes a terminology and information model, machine-readable schema, data sharing conventions, and a reference implementation, each of which is intended to be broadly useful and freely available for community use. VRS was developed by a partnership among national information resource providers, public initiatives, and diagnostic testing laboratories under the auspices of the Global Alliance for Genomics and Health (GA4GH).
Conflict of interest statement
DECLARATION OF INTERESTS H.L.R. is a member of the advisory board for Cell Genomics.
Figures


Similar articles
-
The European Variation Archive: a FAIR resource of genomic variation for all species.Nucleic Acids Res. 2022 Jan 7;50(D1):D1216-D1220. doi: 10.1093/nar/gkab960. Nucleic Acids Res. 2022. PMID: 34718739 Free PMC article.
-
Development and application of a computable genotype model in the GA4GH Variation Representation Specification.Pac Symp Biocomput. 2023;28:383-394. Pac Symp Biocomput. 2023. PMID: 36540993 Free PMC article.
-
ClinGen advancing genomic data-sharing standards as a GA4GH driver project.Hum Mutat. 2018 Nov;39(11):1686-1689. doi: 10.1002/humu.23625. Hum Mutat. 2018. PMID: 30311379 Free PMC article.
-
Federated sharing and processing of genomic datasets for tertiary data analysis.Brief Bioinform. 2021 May 20;22(3):bbaa091. doi: 10.1093/bib/bbaa091. Brief Bioinform. 2021. PMID: 34020536 Review.
-
Assessing the readiness of Turkish health information systems for integrating genetic/genomic patient data: System architecture and available terminologies, legislative, and protection of personal data.Health Policy. 2021 Feb;125(2):203-212. doi: 10.1016/j.healthpol.2020.12.004. Epub 2020 Dec 8. Health Policy. 2021. PMID: 33342546 Review.
Cited by
-
vcfdist: accurately benchmarking phased small variant calls in human genomes.Nat Commun. 2023 Dec 9;14(1):8149. doi: 10.1038/s41467-023-43876-x. Nat Commun. 2023. PMID: 38071244 Free PMC article.
-
SARS-CoV-2 genomic contextual data harmonization: recommendations from a mixed methods analysis of COVID-19 case report forms across Canada.Arch Public Health. 2025 Apr 30;83(1):117. doi: 10.1186/s13690-025-01604-5. Arch Public Health. 2025. PMID: 40307880 Free PMC article.
-
The European Variation Archive: a FAIR resource of genomic variation for all species.Nucleic Acids Res. 2022 Jan 7;50(D1):D1216-D1220. doi: 10.1093/nar/gkab960. Nucleic Acids Res. 2022. PMID: 34718739 Free PMC article.
-
Guidelines for releasing a variant effect predictor.Genome Biol. 2025 Apr 15;26(1):97. doi: 10.1186/s13059-025-03572-z. Genome Biol. 2025. PMID: 40234898 Free PMC article.
-
Candidate targets of copy number deletion events across 17 cancer types.Front Genet. 2023 Jan 16;13:1017657. doi: 10.3389/fgene.2022.1017657. eCollection 2022. Front Genet. 2023. PMID: 36726722 Free PMC article.
References
-
- ENCODE Project Consortium The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. - PubMed
Grants and funding
- R35 HG011949/HG/NHGRI NIH HHS/United States
- U24 HG006834/HG/NHGRI NIH HHS/United States
- K99 HG010157/HG/NHGRI NIH HHS/United States
- U41 HG006834/HG/NHGRI NIH HHS/United States
- R00 HG010157/HG/NHGRI NIH HHS/United States
- U24 HG011025/HG/NHGRI NIH HHS/United States
- U01 CA242954/CA/NCI NIH HHS/United States
- R00 HG007940/HG/NHGRI NIH HHS/United States
- U24 CA237719/CA/NCI NIH HHS/United States
- U54 HG007990/HG/NHGRI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- U41 HG009650/HG/NHGRI NIH HHS/United States
- R35 HG011899/HG/NHGRI NIH HHS/United States

