Carbon starvation, senescence and specific mitochondrial stresses, but not nitrogen starvation and general stresses, are major triggers for mitophagy in Arabidopsis
- PMID: 35311445
- PMCID: PMC9673927
- DOI: 10.1080/15548627.2022.2054039
Carbon starvation, senescence and specific mitochondrial stresses, but not nitrogen starvation and general stresses, are major triggers for mitophagy in Arabidopsis
Abstract
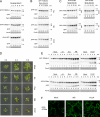
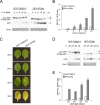
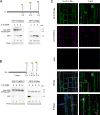
Selective degradation of mitochondria by autophagy (mitophagy) is thought to play an important role in mitochondrial quality control, but our understanding of which conditions induce mitophagy in plants is limited. Here, we developed novel reporter lines to monitor mitophagy in plants and surveyed the rate of mitophagy under a wide range of stresses and developmental conditions. Especially carbon starvation induced by dark-incubation causes a dramatic increase in mitophagy within a few hours, further increasing as dark-induced senescence progresses. Natural senescence was also a strong trigger of mitophagy, peaking when leaf yellowing became prominent. In contrast, nitrogen starvation, a trigger of general autophagy, does not induce strong increases in mitophagy. Similarly, general stresses such as hydrogen peroxide, heat, UV-B and hypoxia did not appear to trigger substantial mitophagy in plants. Additionally, we exposed plants to inhibitors of the mitochondrial electron transport chain, mitochondrial translation and protein import. Although short-term treatments did not induce high mitophagy rates, longer term exposures to uncoupling agent and inhibitors of mitochondrial protein import/translation could clearly increase mitophagic flux. These findings could further be confirmed using confocal microscopy. To validate that mitophagy is mediated by the autophagy pathway, we showed that mitophagic flux is abolished or strongly decreased in atg5/AuTophaGy 5 and atg11 mutants, respectively. Finally, we observed high rates of mitophagy in etiolated seedlings, which remarkably was completely repressed within 6 h after light exposure. In conclusion, we propose that dark-induced carbon starvation, natural senescence and specific mitochondrial stresses are key triggers of mitophagy in plants.Abbreviations: AA: antimycin A; ATG: AuToPhagy related; ConA: concanamycin A; DIS: dark-induced senescence; Dox: doxycycline; FCCP: carbonyl cyanide-p-trifluoromethoxyphenylhydrazone; GFP: green fluorescent protein; IDH1: isocitrate dehydrogenase 1; MB: MitoBlock-6; Mito-GFP: transgenic Arabidopsis line expressing a mitochondrially targeted protein fused to GFP; mtETC: mitochondrial electron transport chain; OXPHOS: oxidative phosphorylation; PQC: protein quality control; TOM20: Translocase of Outer Membrane 20.
Keywords: Arabidopsis; autophagy; mitochondria; mitophagy; plants; senescence.
Conflict of interest statement
No conflicts of interested are reported for this work.
Figures




Similar articles
-
Mitophagy in plants: Emerging regulators of mitochondrial targeting for selective autophagy.J Microsc. 2025 Mar;297(3):325-332. doi: 10.1111/jmi.13267. Epub 2024 Jan 31. J Microsc. 2025. PMID: 38297985 Free PMC article. Review.
-
FRIENDLY is required for efficient dark-induced mitophagy and controlled senescence in Arabidopsis.Free Radic Biol Med. 2023 Aug 1;204:1-7. doi: 10.1016/j.freeradbiomed.2023.04.007. Epub 2023 Apr 19. Free Radic Biol Med. 2023. PMID: 37085125
-
AUTOPHAGY-RELATED11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis.Plant Cell. 2014 Feb;26(2):788-807. doi: 10.1105/tpc.113.120014. Epub 2014 Feb 21. Plant Cell. 2014. PMID: 24563201 Free PMC article.
-
Clearance of damaged mitochondria via mitophagy is important to the protective effect of ischemic preconditioning in kidneys.Autophagy. 2019 Dec;15(12):2142-2162. doi: 10.1080/15548627.2019.1615822. Epub 2019 May 22. Autophagy. 2019. PMID: 31066324 Free PMC article.
-
Regulation of PRKN-independent mitophagy.Autophagy. 2022 Jan;18(1):24-39. doi: 10.1080/15548627.2021.1888244. Epub 2021 Feb 25. Autophagy. 2022. PMID: 33570005 Free PMC article. Review.
Cited by
-
Mitochondrial proteases and their roles in mitophagy in plants, animals, and yeast.Plant Cell Physiol. 2025 Aug 12;66(7):973-990. doi: 10.1093/pcp/pcaf038. Plant Cell Physiol. 2025. PMID: 40269524 Free PMC article. Review.
-
Mitophagy in plants: Emerging regulators of mitochondrial targeting for selective autophagy.J Microsc. 2025 Mar;297(3):325-332. doi: 10.1111/jmi.13267. Epub 2024 Jan 31. J Microsc. 2025. PMID: 38297985 Free PMC article. Review.
-
Vacuolar degradation of plant organelles.Plant Cell. 2024 Sep 3;36(9):3036-3056. doi: 10.1093/plcell/koae128. Plant Cell. 2024. PMID: 38657116 Free PMC article. Review.
-
Transcriptomic Analysis of Three Differentially Senescing Maize (Zea mays L.) Inbred Lines upon Heat Stress.Int J Mol Sci. 2023 Jun 6;24(12):9782. doi: 10.3390/ijms24129782. Int J Mol Sci. 2023. PMID: 37372930 Free PMC article.
-
Autophagy during maize endosperm development dampens oxidative stress and promotes mitochondrial clearance.Plant Physiol. 2023 Sep 22;193(2):1395-1415. doi: 10.1093/plphys/kiad340. Plant Physiol. 2023. PMID: 37335933 Free PMC article.
References
-
- Topf U, Wrobel L, Chacinska A. Chatty mitochondria: keeping balance in cellular protein homeostasis. Trends Cell Biol. 2016;26(8):577–586. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
