Dietary manipulation of the gut microbiome in inflammatory bowel disease patients: Pilot study
- PMID: 35311458
- PMCID: PMC8942410
- DOI: 10.1080/19490976.2022.2046244
Dietary manipulation of the gut microbiome in inflammatory bowel disease patients: Pilot study
Abstract
Diet is a modifiable, noninvasive, inexpensive behavior that is crucial in shaping the intestinal microbiome. A microbiome "imbalance" or dysbiosis in inflammatory bowel disease (IBD) is linked to inflammation. Here, we aim to define the impact of specific foods on bacterial species commonly depleted in patients with IBD to better inform dietary treatment. We performed a single-arm, pre-post intervention trial. After a baseline period, a dietary intervention with the IBD-Anti-Inflammatory Diet (IBD-AID) was initiated. We collected stool and blood samples and assessed dietary intake throughout the study. We applied advanced computational approaches to define and model complex interactions between the foods reported and the microbiome. A dense dataset comprising 553 dietary records and 340 stool samples was obtained from 22 participants. Consumption of prebiotics, probiotics, and beneficial foods correlated with increased abundance of Clostridia and Bacteroides, commonly depleted in IBD cohorts. We further show that specific foods categorized as prebiotics or adverse foods are correlated to levels of cytokines in serum (i.e., GM-CSF, IL-6, IL-8, TNF-alpha) that play a central role in IBD pathogenesis. By using robust predictive analytics, this study represents the first steps to detangle diet-microbiome and diet-immune interactions to inform personalized nutrition for patients suffering from dysbiosis-related IBD.
Keywords: IBD; Microbiome; diet; dysbiosis.
Conflict of interest statement
Beth A. McCormick (BAM) is a coinventor on a patent application (PGT/US 18/42116). She, along with her academic institution, stands to gain financially through potential commercialization outcomes resulting from activities associated with the licensing of that intellectual property. The remaining authors do not have any conflict of interest to disclose.
Figures
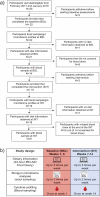

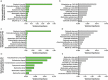



References
-
- Lee JY, Cevallos SA, Byndloss MX, Tiffany CR, Olsan EE, Butler BP, Young BM, Rogers AWL, Nguyen H, Kim K, et al. High-fat diet and antibiotics cooperatively impair mitochondrial bioenergetics to trigger dysbiosis that exacerbates pre-inflammatory bowel disease. Cell Host & Microbe. 2020;28:273–284.e6. doi: 10.1016/j.chom.2020.06.001. - DOI - PMC - PubMed
-
- Frank DN, Robertson CE, Hamm CM, Kpadeh Z, Zhang T, Chen H, Zhu W, Sartor RB, Boedeker EC, Harpaz N, et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflammatory Bowel Diseases. 2011;17(1):179–184. doi: 10.1002/ibd.21339. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
