ATG7-mediated autophagy facilitates embryonic stem cell exit from naive pluripotency and marks commitment to differentiation
- PMID: 35311460
- PMCID: PMC9673953
- DOI: 10.1080/15548627.2022.2055285
ATG7-mediated autophagy facilitates embryonic stem cell exit from naive pluripotency and marks commitment to differentiation
Abstract
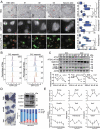
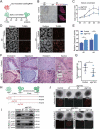
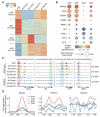
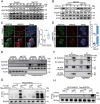
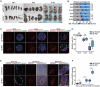
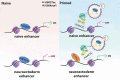
Macroautophagy/autophagy is a conserved cellular mechanism to degrade unneeded cytoplasmic proteins and organelles to recycle their components, and it is critical for embryonic stem cell (ESC) self-renewal and somatic cell reprogramming. Whereas autophagy is essential for early development of embryos, no information exists regarding its functions during the transition from naive-to-primed pluripotency. Here, by using an in vitro transition model of ESCs to epiblast-like cells (EpiLCs), we find that dynamic changes in ATG7-dependent autophagy are critical for the naive-to-primed transition, and are also necessary for germline specification. RNA-seq and ATAC-seq profiling reveal that NANOG acts as a barrier to prevent pluripotency transition, and autophagy-dependent NANOG degradation is important for dismantling the naive pluripotency expression program through decommissioning of naive-associated active enhancers. Mechanistically, we found that autophagy receptor protein SQSTM1/p62 translocated into the nucleus during the pluripotency transition period and is preferentially associated with K63 ubiquitinated NANOG for selective protein degradation. In vivo, loss of autophagy by ATG7 depletion disrupts peri-implantation development and causes increased chromatin association of NANOG, which affects neuronal differentiation by competitively binding to OTX2-specific neuroectodermal development-associated regions. Taken together, our findings reveal that autophagy-dependent degradation of NANOG plays a critical role in regulating exit from the naive state and marks distinct cell fate allocation during lineage specification.Abbreviations: 3-MA: 3-methyladenine; EpiLC: epiblast-like cell; ESC: embryonic stem cell; PGC: primordial germ cell.
Keywords: ATG7; NANOG; autophagy; naive-to-primed transition; peri-implantation development.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


Similar articles
-
Loss of the Otx2-Binding Site in the Nanog Promoter Affects the Integrity of Embryonic Stem Cell Subtypes and Specification of Inner Cell Mass-Derived Epiblast.Cell Rep. 2016 Jun 21;15(12):2651-64. doi: 10.1016/j.celrep.2016.05.041. Epub 2016 Jun 9. Cell Rep. 2016. PMID: 27292645
-
Foxd3 Promotes Exit from Naive Pluripotency through Enhancer Decommissioning and Inhibits Germline Specification.Cell Stem Cell. 2016 Jan 7;18(1):118-33. doi: 10.1016/j.stem.2015.09.010. Cell Stem Cell. 2016. PMID: 26748758 Free PMC article.
-
NANOG alone induces germ cells in primed epiblast in vitro by activation of enhancers.Nature. 2016 Jan 21;529(7586):403-407. doi: 10.1038/nature16480. Epub 2016 Jan 11. Nature. 2016. PMID: 26751055 Free PMC article.
-
Generating primed pluripotent epiblast stem cells: A methodology chapter.Curr Top Dev Biol. 2020;138:139-174. doi: 10.1016/bs.ctdb.2020.01.005. Epub 2020 Feb 27. Curr Top Dev Biol. 2020. PMID: 32220296 Free PMC article. Review.
-
Switching on pluripotency: a perspective on the biological requirement of Nanog.Philos Trans R Soc Lond B Biol Sci. 2011 Aug 12;366(1575):2222-9. doi: 10.1098/rstb.2011.0003. Philos Trans R Soc Lond B Biol Sci. 2011. PMID: 21727127 Free PMC article. Review.
Cited by
-
Autophagy-mediated ID1 turnover dictates chemo-resistant fate in ovarian cancer stem cells.J Exp Clin Cancer Res. 2024 Aug 10;43(1):222. doi: 10.1186/s13046-024-03147-z. J Exp Clin Cancer Res. 2024. PMID: 39123206 Free PMC article.
-
Autophagy regulation combined with stem cell therapy for treatment of spinal cord injury.Neural Regen Res. 2023 Aug;18(8):1629-1636. doi: 10.4103/1673-5374.363189. Neural Regen Res. 2023. PMID: 36751772 Free PMC article. Review.
-
Autophagy accompanying the developmental process of male germline stem cells.Cell Tissue Res. 2024 Oct;398(1):1-14. doi: 10.1007/s00441-024-03910-w. Epub 2024 Aug 14. Cell Tissue Res. 2024. PMID: 39141056 Review.
-
Protective Effect of Photobiomodulation against Hydrogen Peroxide-Induced Oxidative Damage by Promoting Autophagy through Inhibition of PI3K/AKT/mTOR Pathway in MC3T3-E1 Cells.Oxid Med Cell Longev. 2022 Nov 22;2022:7223353. doi: 10.1155/2022/7223353. eCollection 2022. Oxid Med Cell Longev. 2022. PMID: 36457727 Free PMC article.
-
Dynamic Interplay Between Autophagy and Oxidative Stress in Stem Cells: Implications for Regenerative Medicine.Antioxidants (Basel). 2025 Jun 6;14(6):691. doi: 10.3390/antiox14060691. Antioxidants (Basel). 2025. PMID: 40563323 Free PMC article. Review.
References
-
- Hackett JA, Surani MA.. Regulatory principles of pluripotency: from the ground state up. Cell Stem Cell. 2014;15(4):416–430. - PubMed
-
- Kojima Y, Kaufman-Francis K, Studdert JB, et al. The transcriptional and functional properties of mouse epiblast stem cells resemble the anterior primitive streak. Cell Stem Cell. 2014;14(1):107–120. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
