Genome-Wide Assessment of Stress-Associated Genes in Bifidobacteria
- PMID: 35311508
- PMCID: PMC9004370
- DOI: 10.1128/aem.02251-21
Genome-Wide Assessment of Stress-Associated Genes in Bifidobacteria
Abstract
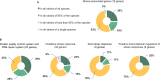
Over the last decade, the genomes of several Bifidobacterium strains have been sequenced, delivering valuable insights into their genetic makeup. However, bifidobacterial genomes have not yet been systematically mined for genes associated with stress response functions and their regulation. In this work, a list of 76 genes related to stress response in bifidobacteria was compiled from previous studies. The prevalence of the genes was evaluated among the genome sequences of 171 Bifidobacterium strains. Although genes of the protein quality control and DNA repair systems appeared to be highly conserved, genome-wide in silico screening for consensus sequences of putative regulators suggested that the regulation of these systems differs among phylogenetic groups. Homologs of multiple oxidative stress-associated genes are shared across species, albeit at low sequence similarity. Bee isolates were confirmed to harbor unique genetic features linked to oxygen tolerance. Moreover, most studied Bifidobacterium adolescentis and all Bifidobacterium angulatum strains lacked a set of reactive oxygen species-detoxifying enzymes, which might explain their high sensitivity to oxygen. Furthermore, the presence of some putative transcriptional regulators of stress responses was found to vary across species and strains, indicating that different regulation strategies of stress-associated gene transcription contribute to the diverse stress tolerance. The presented stress response gene profiles of Bifidobacterium strains provide a valuable knowledge base for guiding future studies by enabling hypothesis generation and the identification of key genes for further analyses. IMPORTANCE Bifidobacteria are Gram-positive bacteria that naturally inhabit diverse ecological niches, including the gastrointestinal tract of humans and animals. Strains of the genus Bifidobacterium are widely used as probiotics, since they have been associated with health benefits. In the course of their production and administration, probiotic bifidobacteria are exposed to several stressors that can challenge their survival. The stress tolerance of probiotic bifidobacteria is, therefore, an important selection criterion for their commercial application, since strains must maintain their viability to exert their beneficial health effects. As the ability to cope with stressors varies among Bifidobacterium strains, comprehensive understanding of the underlying stress physiology is required for enabling knowledge-driven strain selection and optimization of industrial-scale production processes.
Keywords: bifidobacteria; genomics; stress response.
Conflict of interest statement
The authors declare a conflict of interest. M.S., K.J., T.V., and A.A.Z. were employed by Chr. Hansen A/S, a global supplier of probiotics, at the time of writing the manuscript. The authors' views presented in this manuscript, however, are solely based on scientific grounds and do not reflect the commercial interests of the employer.
Figures


Similar articles
-
Stress Response in Bifidobacteria.Microbiol Mol Biol Rev. 2022 Dec 21;86(4):e0017021. doi: 10.1128/mmbr.00170-21. Epub 2022 Nov 14. Microbiol Mol Biol Rev. 2022. PMID: 36374074 Free PMC article. Review.
-
Novel Insights into the Molecular Mechanisms Underlying Robustness and Stability in Probiotic Bifidobacteria.Appl Environ Microbiol. 2023 Mar 29;89(3):e0008223. doi: 10.1128/aem.00082-23. Epub 2023 Feb 21. Appl Environ Microbiol. 2023. PMID: 36802222 Free PMC article.
-
Bifidobacterium xylocopae sp. nov. and Bifidobacterium aemilianum sp. nov., from the carpenter bee (Xylocopa violacea) digestive tract.Syst Appl Microbiol. 2019 Mar;42(2):205-216. doi: 10.1016/j.syapm.2018.11.005. Epub 2018 Nov 29. Syst Appl Microbiol. 2019. PMID: 30551956
-
Probiotic characters of Bifidobacterium and Lactobacillus are a result of the ongoing gene acquisition and genome minimization evolutionary trends.Microb Pathog. 2017 Oct;111:118-131. doi: 10.1016/j.micpath.2017.08.021. Epub 2017 Aug 18. Microb Pathog. 2017. PMID: 28826768 Review.
-
Identification and characterization of WhiB-like family proteins of the Bifidobacterium genus.Anaerobe. 2012 Aug;18(4):421-9. doi: 10.1016/j.anaerobe.2012.04.011. Epub 2012 May 15. Anaerobe. 2012. PMID: 22609519
Cited by
-
The influence of the gut-brain axis on anxiety and depression: A review of the literature on the use of probiotics.J Tradit Complement Med. 2024 Mar 21;14(3):237-255. doi: 10.1016/j.jtcme.2024.03.011. eCollection 2024 May. J Tradit Complement Med. 2024. PMID: 38707924 Free PMC article. Review.
-
Exploring Bifidobacterium species community and functional variations with human gut microbiome structure and health beyond infancy.Microbiome Res Rep. 2023 Mar 31;2(2):9. doi: 10.20517/mrr.2023.01. eCollection 2023. Microbiome Res Rep. 2023. PMID: 38047280 Free PMC article.
-
Deciphering probiotic potential: a comprehensive guide to probiogenomic analyses.Future Microbiol. 2025 May-Jun;20(7-9):611-622. doi: 10.1080/17460913.2025.2492472. Epub 2025 Apr 14. Future Microbiol. 2025. PMID: 40227157 Review.
-
Anti-Inflammatory Function Analysis of Lacticaseibacillus rhamnosus CP-1 Strain Based on Whole-Genome Sequencing.BioTech (Basel). 2025 Jun 7;14(2):47. doi: 10.3390/biotech14020047. BioTech (Basel). 2025. PMID: 40558396 Free PMC article.
-
Stress Response in Bifidobacteria.Microbiol Mol Biol Rev. 2022 Dec 21;86(4):e0017021. doi: 10.1128/mmbr.00170-21. Epub 2022 Nov 14. Microbiol Mol Biol Rev. 2022. PMID: 36374074 Free PMC article. Review.
References
-
- Rosberg-Cody E, Liavonchanka A, Göbel C, Ross RP, O'Sullivan O, Fitzgerald GF, Feussner I, Stanton C. 2011. Myosin-cross-reactive antigen (MCRA) protein from Bifidobacterium breve is a FAD-dependent fatty acid hydratase which has a function in stress protection. BMC Biochem 12:9. 10.1186/1471-2091-12-9. - DOI - PMC - PubMed
-
- Ventura M, Zink R, Fitzgerald GF, Van Sinderen D. 2005. Gene structure and transcriptional organization of the dnaK operon of Bifidobacterium breve UCC 2003 and application of the operon in bifidobacterial tracing. Appl Environ Microbiol 71:487–500. 10.1128/AEM.71.1.487-500.2005. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

