Microbial Sources of Exocellular DNA in the Ocean
- PMID: 35311515
- PMCID: PMC9004351
- DOI: 10.1128/aem.02093-21
Microbial Sources of Exocellular DNA in the Ocean
Abstract
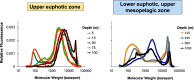
Exocellular DNA is operationally defined as the fraction of the total DNA pool that passes through a membrane filter (0.1 μm). It is composed of DNA-containing vesicles, viruses, and free DNA and is ubiquitous in all aquatic systems, although the sources, sinks, and ecological consequences are largely unknown. Using a method that provides separation of these three fractions, we compared open ocean depth profiles of DNA associated with each fraction. Pelagibacter-like DNA dominated the vesicle fractions for all samples examined over a depth range of 75 to 500 m. Viral DNA consisted predominantly of myovirus-like and podovirus-like DNA and contained the highest proportion of unannotated sequences. Euphotic zone free DNA (75 to 125 m) contained primarily bacterial and viral sequences, with bacteria dominating samples from the mesopelagic zone (500 to 1,000 m). A high proportion of mesopelagic zone free DNA sequences appeared to originate from surface waters, including a large amount of DNA contributed by high-light Prochlorococcus ecotypes. Throughout the water column, but especially in the mesopelagic zone, the composition of free DNA sequences was not always reflective of cooccurring microbial communities that inhabit the same sampling depth. These results reveal the composition of free DNA in different regions of the water column (euphotic and mesopelagic zones), with implications for dissolved organic matter cycling and export (by way of sinking particles and/or migratory zooplankton) as a delivery mechanism. IMPORTANCE With advances in metagenomic sequencing, the microbial composition of diverse environmental systems has been investigated, providing new perspectives on potential ecological dynamics and dimensions for experimental investigations. Here, we characterized exocellular free DNA via metagenomics, using a newly developed method that separates free DNA from cells, viruses, and vesicles, and facilitated the independent characterization of each fraction. The fate of this free DNA has both ecological consequences as a nutrient (N and P) source and potential evolutionary consequences as a source of genetic transformation. Here, we document different microbial sources of free DNA at the surface (0 to 200 m) versus depths of 250 to 1,000 m, suggesting that distinct free DNA production mechanisms may be present throughout the oligotrophic water column. Examining microbial processes through the lens of exocellular DNA provides insights into the production of labile dissolved organic matter (i.e., free DNA) at the surface (likely by viral lysis) and processes that influence the fate of sinking, surface-derived organic matter.
Keywords: bacteriophage; bacterioplankton; eDNA; exocellular DNA; free DNA; metagenomics; microbial ecology; microbial oceanography; open ocean; vesicle.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Holm-Hansen O, Sutcliffe WH, Jr, Sharp J. 1968. Measurement of deoxyribonucleic acid in the ocean and its ecological significance. Limnol Oceanogr 13:507–514. 10.4319/lo.1968.13.3.0507. - DOI
-
- Winn CD, Karl DM. 1986. Diel nucleic acid synthesis and particulate DNA concentrations: conflicts with division rate estimates by DNA accumulation. Limnol Oceanogr 31:637–645. 10.4319/lo.1986.31.3.0637. - DOI
-
- DeFlaun MF, Paul JH, Jeffrey WH. 1987. Distribution and molecular weight of dissolved DNA in subtropical estuarine and oceanic environments. Mar Ecol Prog Ser 38:65–73. 10.3354/meps038065. - DOI
-
- Dell’Anno A, Marrale D, Pusceddu A, Fabiano M, Danovaro R. 1999. Particulate nucleic acid dynamics in a highly oligotrophic system: the Cretan Sea (Eastern Mediterranean). Mar Ecol Prog Ser 186:19–30. 10.3354/meps186019. - DOI
-
- Karl DM, Bailiff MD. 1989. The measurement and distribution of dissolved nucleic acids in aquatic environments. Limnol Oceanogr 34:543–558. 10.4319/lo.1989.34.3.0543. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

