Proximal Binding Pocket Arg717 Substitutions in Escherichia coli AcrB Cause Clinically Relevant Divergencies in Resistance Profiles
- PMID: 35311521
- PMCID: PMC9017336
- DOI: 10.1128/aac.02392-21
Proximal Binding Pocket Arg717 Substitutions in Escherichia coli AcrB Cause Clinically Relevant Divergencies in Resistance Profiles
Abstract
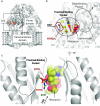
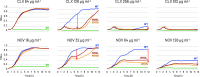
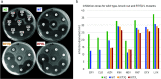
Recent mutations in RND efflux pumps in clinical strains have further increased multidrug resistance. We show that R717L and R717Q substitutions (found in azithromycin-resistant Salmonella enterica spp.) in the Escherichia coli efflux pump AcrB dramatically increase macrolide, as well as fluoroquinolone, resistance. On the other hand, cells became more susceptible to novobiocin and β-lactam cloxacillin. We urge the control of, and adjustments to, treatments with antibiotics and the need for novel antibiotics and efflux pump inhibitors.
Keywords: AcrB; RND; antimicrobial resistance; fluoroquinolones; macrolides; multidrug resistance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Metabolomics Reveal Potential Natural Substrates of AcrB in Escherichia coli and Salmonella enterica Serovar Typhimurium.mBio. 2021 Mar 30;12(2):e00109-21. doi: 10.1128/mBio.00109-21. mBio. 2021. PMID: 33785633 Free PMC article.
-
Kinetic analysis of the inhibition of the drug efflux protein AcrB using surface plasmon resonance.Biochim Biophys Acta Biomembr. 2018 Apr;1860(4):878-886. doi: 10.1016/j.bbamem.2017.08.024. Epub 2017 Sep 8. Biochim Biophys Acta Biomembr. 2018. PMID: 28890187
-
Reversal of the Drug Binding Pocket Defects of the AcrB Multidrug Efflux Pump Protein of Escherichia coli.J Bacteriol. 2015 Oct;197(20):3255-64. doi: 10.1128/JB.00547-15. Epub 2015 Aug 3. J Bacteriol. 2015. PMID: 26240069 Free PMC article.
-
AcrB: a mean, keen, drug efflux machine.Ann N Y Acad Sci. 2020 Jan;1459(1):38-68. doi: 10.1111/nyas.14239. Epub 2019 Oct 6. Ann N Y Acad Sci. 2020. PMID: 31588569 Review.
-
The AcrB efflux pump: conformational cycling and peristalsis lead to multidrug resistance.Curr Drug Targets. 2008 Sep;9(9):729-49. doi: 10.2174/138945008785747789. Curr Drug Targets. 2008. PMID: 18781920 Review.
Cited by
-
Comparative reassessment of AcrB efflux inhibitors reveals differential impact of specific pump mutations on the activity of potent compounds.Microbiol Spectr. 2024 Feb 6;12(2):e0304523. doi: 10.1128/spectrum.03045-23. Epub 2024 Jan 3. Microbiol Spectr. 2024. PMID: 38170977 Free PMC article.
-
Structural and functional analysis of the Mycobacterium tuberculosis MmpS5L5 efflux pump presages a pathway to increased bedaquiline resistance.bioRxiv [Preprint]. 2025 Jun 24:2025.06.24.661325. doi: 10.1101/2025.06.24.661325. bioRxiv. 2025. PMID: 40667120 Free PMC article. Preprint.
-
Structural and functional diversity of Resistance-Nodulation-Division (RND) efflux pump transporters with implications for antimicrobial resistance.Microbiol Mol Biol Rev. 2024 Sep 26;88(3):e0008923. doi: 10.1128/mmbr.00089-23. Epub 2024 Sep 5. Microbiol Mol Biol Rev. 2024. PMID: 39235227 Review.
-
Functionally distinct mutations within AcrB underpin antibiotic resistance in different lifestyles.NPJ Antimicrob Resist. 2023;1(1):2. doi: 10.1038/s44259-023-00001-8. Epub 2023 May 10. NPJ Antimicrob Resist. 2023. PMID: 38686215 Free PMC article.
-
RND pumps across the genus Acinetobacter: AdeIJK is the universal efflux pump.Microb Genom. 2023 Mar;9(3):mgen000964. doi: 10.1099/mgen.0.000964. Microb Genom. 2023. PMID: 36995182 Free PMC article.
References
-
- World Health Organization. 2015. Global action plan on antimicrobial resistance. World Health Organization: Geneva, Switzerland. https://www.who.int/publications/i/item/9789241509763. Accessed 01 December 2021.
-
- World Health Organization. 2014. Antimicrobial resistance global report on surveillance: 2014 summary. World Health Organization: Geneva, Switzerland. https://apps.who.int/iris/handle/10665/112642. Accessed 01 December 2021.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

