Analysis of mRNA Decay Intermediates in Bacillus subtilis 3' Exoribonuclease and RNA Helicase Mutant Strains
- PMID: 35311531
- PMCID: PMC9040804
- DOI: 10.1128/mbio.00400-22
Analysis of mRNA Decay Intermediates in Bacillus subtilis 3' Exoribonuclease and RNA Helicase Mutant Strains
Abstract
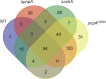
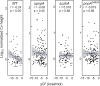
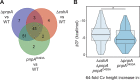
The Bacillus subtilis genome encodes four 3' exoribonucleases: polynucleotide phosphorylase (PNPase), RNase R, RNase PH, and YhaM. Previous work showed that PNPase, encoded by the pnpA gene, is the major 3' exonuclease involved in mRNA turnover; in a pnpA deletion strain, numerous mRNA decay intermediates accumulate. Whether B. subtilis mRNA decay occurs in the context of a degradosome complex is controversial. In this study, global mapping of mRNA decay intermediate 3' ends within coding sequences was performed in strains that were either deleted for or had an inactivating point mutation in the pnpA gene. The patterns of 3'-end accumulation in these strains were highly similar, which may have implications for the role of a degradosome in mRNA decay. A comparison with mapped 3' ends in a strain lacking CshA, the major RNA helicase, indicated that many mRNAs require both PNPase and CshA for efficient decay. Transcriptome sequencing (RNA-seq) analysis of strains lacking RNase R suggested that this enzyme did not play a major role in mRNA turnover in the wild-type strain. Strains were constructed that contained only one of the four known 3' exoribonucleases. When RNase R was the only 3' exonuclease present, it was able to degrade a model mRNA efficiently, showing processive decay even through a strong stem-loop structure that inhibits PNPase processivity. Strains containing only RNase PH or only YhaM were also insensitive to this RNA secondary structure, suggesting the existence of another, as-yet-unidentified, 3' exoribonuclease. IMPORTANCE The ability to rapidly change bacterial gene expression programs in response to environmental conditions is highly dependent on the efficient turnover of mRNA. While much is known about the regulation of gene expression at the transcriptional and translational levels, much less is known about the intermediate step of mRNA decay. Here, we mapped the 3' ends of mRNA decay intermediates in strains that were missing the major 3' exoribonuclease PNPase or the RNA helicase CshA. We also assessed the roles of three other B. subtilis 3' exonucleases in the mRNA decay process. The data confirm the primary role of PNPase in mRNA turnover and suggest the involvement of one or more unknown RNases.
Keywords: 3′ exoribonucleases; Bacillus subtilis; PNPase; RNA helicase; Term-seq; mRNA decay; transcriptomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

