Interactions and Stability of Gut Microbiota in Zebrafish Increase with Host Development
- PMID: 35311546
- PMCID: PMC9045336
- DOI: 10.1128/spectrum.01696-21
Interactions and Stability of Gut Microbiota in Zebrafish Increase with Host Development
Abstract
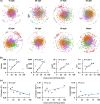
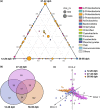
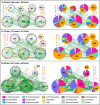
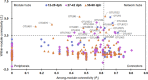
Understanding interactions within the gut microbiome and its stability are of critical importance for deciphering ecological issues within the gut ecosystem. Recent studies indicate that long-term instability of gut microbiota is associated with human diseases, and recovery of stability is helpful in the return to health. However, much less is known about such topics in fish, which encompass nearly half of all vertebrate diversity. Here, we examined the assembly and succession of gut microbiota in more than 550 zebrafish, and evaluated the variations of microbial interactions and stability across fish development from larva to adult using molecular ecological network analysis. We found that microbial interactions and stability in the fish gut ecosystem generally increased with host development. This could be attributed to the development of the zebrafish immune system, the increasing amount of space available for microbial colonization within the gut, and the greater stability of nutrients available for the colonized microbiota in adult zebrafish. Moreover, the potential keystone taxa, even those with relatively low abundances, played important roles in affecting the microbial interactions and stability. These findings indicate that regulating rare keystone taxa in adult fish may have great potential in gut microbial management to maintain gut ecosystem stability, which could also provide references for managing gut microbiota in humans and other animals. IMPORTANCE Understanding gut microbial stability and the underlying mechanisms is an important but largely ignored ecological issue in vertebrate fish. Here, using a zebrafish model and network analysis of the gut microbiota we found that microbial interactions and stability in the gut ecosystem increase with fish development. This finding has important implications for microbial management to maintain gut homeostasis and provide better gut ecosystem services for the host. First, future studies should always consider using fish of different age groups to gain a full understanding of gut microbial networks. Second, management of the keystone taxa, even those that are only present at a low abundance, during the adult stage may be a viable pathway to maintain gut ecosystem stability. This study greatly expands our current knowledge regarding gut ecosystem stability in terms of ecological networks affected by fish development, and also highlights potential directions for gut microbial management in humans and other animals.
Keywords: ecosystem stability; gut microbiota; keystone taxa; microbial interactions; zebrafish.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Variable phylosymbiosis and cophylogeny patterns in wild fish gut microbiota of a large subtropical river.mSphere. 2025 Apr 29;10(4):e0098224. doi: 10.1128/msphere.00982-24. Epub 2025 Mar 28. mSphere. 2025. PMID: 40152595 Free PMC article.
-
Resistance and Resilience of Fish Gut Microbiota to Silver Nanoparticles.mSystems. 2021 Oct 26;6(5):e0063021. doi: 10.1128/mSystems.00630-21. Epub 2021 Sep 14. mSystems. 2021. PMID: 34519523 Free PMC article.
-
Environmental effects of nanoparticles on the ecological succession of gut microbiota across zebrafish development.Sci Total Environ. 2022 Feb 1;806(Pt 4):150963. doi: 10.1016/j.scitotenv.2021.150963. Epub 2021 Oct 15. Sci Total Environ. 2022. PMID: 34656599
-
Feed, Microbiota, and Gut Immunity: Using the Zebrafish Model to Understand Fish Health.Front Immunol. 2020 Feb 5;11:114. doi: 10.3389/fimmu.2020.00114. eCollection 2020. Front Immunol. 2020. PMID: 32117265 Free PMC article. Review.
-
Interwoven processes in fish development: microbial community succession and immune maturation.PeerJ. 2024 Mar 27;12:e17051. doi: 10.7717/peerj.17051. eCollection 2024. PeerJ. 2024. PMID: 38560465 Free PMC article. Review.
Cited by
-
Microbial Interactions in Rearing Systems for Marine Fish Larvae.Microorganisms. 2025 Feb 27;13(3):539. doi: 10.3390/microorganisms13030539. Microorganisms. 2025. PMID: 40142430 Free PMC article. Review.
-
Early Succession of Community Structures and Biotic Interactions of Gut Microbes in Eriocheir sinensis Megalopa after Desalination.Microorganisms. 2024 Mar 11;12(3):560. doi: 10.3390/microorganisms12030560. Microorganisms. 2024. PMID: 38543611 Free PMC article.
-
Variation in the gut microbiota during the early developmental stages of common carp (Cyprinus carpio L.) and its correlation with feed and pond water microflora.BMC Vet Res. 2024 Oct 11;20(1):464. doi: 10.1186/s12917-024-04321-3. BMC Vet Res. 2024. PMID: 39394135 Free PMC article.
-
Probiotic supplementation during pregnancy alters gut microbial networks of pregnant women and infants.Front Microbiol. 2022 Dec 1;13:1042846. doi: 10.3389/fmicb.2022.1042846. eCollection 2022. Front Microbiol. 2022. PMID: 36532501 Free PMC article.
-
Red Ginseng Dietary Fiber Shows Prebiotic Potential by Modulating Gut Microbiota in Dogs.Microbiol Spectr. 2023 Aug 17;11(4):e0094923. doi: 10.1128/spectrum.00949-23. Epub 2023 Jun 27. Microbiol Spectr. 2023. PMID: 37367492 Free PMC article.
References
-
- Xiao F, Zhu W, Yu Y, He Z, Wu B, Wang C, Shu L, Li X, Yin H, Wang J, Juneau P, Zheng X, Wu Y, Li J, Chen X, Hou D, Huang Z, He J, Xu G, Xie L, Huang J, Yan Q. 2021. Host development overwhelms environmental dispersal in governing the ecological succession of zebrafish gut microbiota. NPJ Biofilms Microbiomes 7:5. doi:10.1038/s41522-020-00176-2. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

