Presence of Natural Killer B Cells in Simian Immunodeficiency Virus-Infected Colon That Have Properties and Functions Similar to Those of Natural Killer Cells and B Cells but Are a Distinct Cell Population
- PMID: 35311549
- PMCID: PMC9006943
- DOI: 10.1128/jvi.00235-22
Presence of Natural Killer B Cells in Simian Immunodeficiency Virus-Infected Colon That Have Properties and Functions Similar to Those of Natural Killer Cells and B Cells but Are a Distinct Cell Population
Abstract
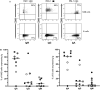
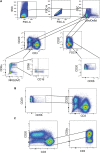
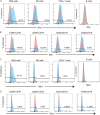
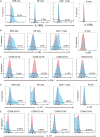
Here, we report the appearance of natural killer B (NKB) cells within the colon during simian immunodeficiency virus (SIV) infection of susceptible monkeys. Using RNA sequencing (RNAseq) and flow cytometry, we show that NKB cells are unique cells with features and functions of both NK and B cells. NKB cells express receptors and ligands found on B cells that are important for (i) antigen presentation; (ii) activities associated with class switching, affinity maturation, and B-cell memory formation in secondary lymphoid follicles; and (iii) antigen recognition. The predominant immunoglobulins (Igs) expressed on NKB cells are IgA, although NKB cells can express surface IgM and IgG. There is dominant lambda expression over the kappa light chain characteristic of mucosal B cells. In addition to B-cell aspects, NKB cells express NK cell activation receptors and Fas ligand. We show in this study that NKB cells express perforin and granzymes and lyse cells in a lytic assay. In addition to NK cell cytolytic function, NKB cells also produce the inflammatory cytokines interferon gamma, tumor necrosis factor alpha, and interleukin-18 (IL-18). Finally, we noted the increased capacity of NKB cells to proliferate compared to NK cells and CD8+ T cells from the SIV-infected colon. The increased proliferation and inflammatory cytokine production may be related to the relatively high expression levels of IL-15 receptor beta, IL-7 receptor, IL-18 receptor, and 41BB relative to the same receptors on CD8 and NK cells. The properties of NKB cells may point to their role in the enhanced inflammation observed in the SIV-infected gut. IMPORTANCE There is low-level but significant mucosal inflammation in the gastrointestinal tract secondary to human immunodeficiency virus (HIV) infection that has long-term consequences for the infected host. This inflammation most likely originates from the immune response that appears as a consequence of HIV. Here, we show in an animal model of HIV that the chronically SIV-infected gut contains cytotoxic natural killer B cells that produce inflammatory cytokines and proliferate during infection.
Keywords: B cells; cytokines; cytotoxic; gastrointestinal; gut inflammation; human immunodeficiency virus; immunoglobulin A; mucosal immunity; natural killer cells; simian immunodeficiency virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







Similar articles
-
Presence of Inflammatory Group I and III Innate Lymphoid Cells in the Colon of Simian Immunodeficiency Virus-Infected Rhesus Macaques.J Virol. 2020 Apr 16;94(9):e01914-19. doi: 10.1128/JVI.01914-19. Print 2020 Apr 16. J Virol. 2020. PMID: 32051277 Free PMC article.
-
Functional Perturbation of Mucosal Group 3 Innate Lymphoid and Natural Killer Cells in Simian-Human Immunodeficiency Virus/Simian Immunodeficiency Virus-Infected Infant Rhesus Macaques.J Virol. 2020 Feb 14;94(5):e01644-19. doi: 10.1128/JVI.01644-19. Print 2020 Feb 14. J Virol. 2020. PMID: 31801861 Free PMC article.
-
Interleukin-21 administration to rhesus macaques chronically infected with simian immunodeficiency virus increases cytotoxic effector molecules in T cells and NK cells and enhances B cell function without increasing immune activation or viral replication.Vaccine. 2011 Nov 15;29(49):9229-38. doi: 10.1016/j.vaccine.2011.09.118. Epub 2011 Oct 11. Vaccine. 2011. PMID: 21996099 Free PMC article.
-
Role of follicular homing natural killer cells in HIV infection.Curr Opin HIV AIDS. 2025 Mar 1;20(2):154-158. doi: 10.1097/COH.0000000000000916. Epub 2025 Jan 2. Curr Opin HIV AIDS. 2025. PMID: 39773847 Review.
-
Monkeying Around: Using Non-human Primate Models to Study NK Cell Biology in HIV Infections.Front Immunol. 2019 May 22;10:1124. doi: 10.3389/fimmu.2019.01124. eCollection 2019. Front Immunol. 2019. PMID: 31191520 Free PMC article. Review.
Cited by
-
NKB cells: A double-edged sword against inflammatory diseases.Front Immunol. 2022 Nov 3;13:972435. doi: 10.3389/fimmu.2022.972435. eCollection 2022. Front Immunol. 2022. PMID: 36405684 Free PMC article. Review.
-
Noncanonical B Cells: Characteristics of Uncharacteristic B Cells.J Immunol. 2023 Nov 1;211(9):1257-1265. doi: 10.4049/jimmunol.2200944. J Immunol. 2023. PMID: 37844278 Free PMC article. Review.
-
Virus Protein-Specific Immune Responses in Selective Depletion of Lymphocyte Populations Using Monoclonal Antibodies in Bolivian Squirrel Monkeys (Saimiri boliviensis boliviensisv).Viral Immunol. 2025 Jan-Feb;38(1):12-22. doi: 10.1089/vim.2024.0080. Epub 2025 Jan 2. Viral Immunol. 2025. PMID: 39745246
-
Natural killer-like B cells are a distinct but infrequent innate immune cell subset modulated by SIV infection of rhesus macaques.PLoS Pathog. 2024 May 13;20(5):e1012223. doi: 10.1371/journal.ppat.1012223. eCollection 2024 May. PLoS Pathog. 2024. PMID: 38739675 Free PMC article.
-
Regulatory B Cells Expressing Granzyme B from Tolerant Renal Transplant Patients: Highly Differentiated B Cells with a Unique Pathway with a Specific Regulatory Profile and Strong Interactions with Immune System Cells.Cells. 2024 Jul 31;13(15):1287. doi: 10.3390/cells13151287. Cells. 2024. PMID: 39120317 Free PMC article.
References
-
- Macdonald TT. 2008. The gut is still the biggest lymphoid organ in the body. Mucosal Immunol 1:246–247.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

