Ribitol-Containing Wall Teichoic Acid of Tetragenococcus halophilus Is Targeted by Bacteriophage phiWJ7 as a Binding Receptor
- PMID: 35311554
- PMCID: PMC9045211
- DOI: 10.1128/spectrum.00336-22
Ribitol-Containing Wall Teichoic Acid of Tetragenococcus halophilus Is Targeted by Bacteriophage phiWJ7 as a Binding Receptor
Abstract
Tetragenococcus halophilus, a halophilic lactic acid bacterium, is used in the fermentation process of soy sauce manufacturing. For many years, bacteriophage infections of T. halophilus have been a major industrial problem that causes fermentation failure. However, studies focusing on the mechanisms of tetragenococcal host-phage interactions are not sufficient. In this study, we generated two phage-insensitive derivatives from the parental strain T. halophilus WJ7, which is susceptible to the virulent phage phiWJ7. Whole-genome sequencing of the derivatives revealed that insertion sequences were transposed into a gene encoding poly(ribitol phosphate) polymerase (TarL) in both derivatives. TarL is responsible for the biosynthesis of ribitol-containing wall teichoic acid, and WJ7 was confirmed to contain ribitol in extracted wall teichoic acid, but the derivative was not. Cell walls of WJ7 irreversibly adsorbed phiWJ7, but those of the phage-insensitive derivatives did not. Additionally, 25 phiWJ7-insensitive derivatives were obtained, and they showed mutations not only in tarL but also in tarI and tarJ, which are responsible for the synthesis of CDP-ribitol. These results indicate that phiWJ7 targets the ribitol-containing wall teichoic acid of host cells as a binding receptor. IMPORTANCE Information about the mechanisms of host-phage interactions is required for the development of efficient strategies against bacteriophage infections. Here, we identified the ribitol-containing wall teichoic acid as a host receptor indispensable for bacteriophage infection. The complete genome sequence of tetragenococcal phage phiWJ7 belonging to the family Rountreeviridae is also provided here. This study could become the foundation for a better understanding of host-phage interactions of tetragenococci.
Keywords: Tetragenococcus halophilus; bacteriophage; host receptor; wall teichoic acid.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
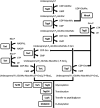
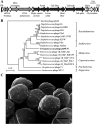

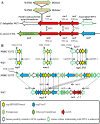


References
-
- Kuda T, Izawa Y, Ishii S, Takahashi H, Torido Y, Kimura B. 2012. Suppressive effect of Tetragenococcus halophilus, isolated from fish-nukazuke, on histamine accumulation in salted and fermented fish. Food Chem 130:569–574. doi: 10.1016/j.foodchem.2011.07.074. - DOI
-
- Uchida K, Kanbe C. 1993. Occurrence of bacteriophages lytic for Pediococcus halophilus, a halophilic lactic-acid bacterium, in soy sauce fermentation. J Gen Appl Microbiol 39:429–437. doi: 10.2323/jgam.39.429. - DOI
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

