The Rcs System Contributes to the Motility Defects of the Twin-Arginine Translocation System Mutant of Extraintestinal Pathogenic Escherichia coli
- PMID: 35311558
- PMCID: PMC9017377
- DOI: 10.1128/jb.00612-21
The Rcs System Contributes to the Motility Defects of the Twin-Arginine Translocation System Mutant of Extraintestinal Pathogenic Escherichia coli
Abstract
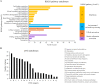
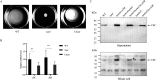
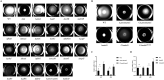
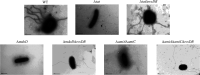
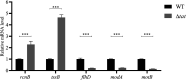
Flagellum-mediated bacterial motility is important for bacteria to take up nutrients, adapt to environmental changes, and establish infection. The twin-arginine translocation system (Tat) is an important protein export system, playing a critical role in bacterial physiology and pathogenesis. It has been observed for a long time that the Tat system is critical for bacterial motility. However, the underlying mechanism remains unrevealed. In this study, a comparative transcriptomics analysis was performed with extraintestinal pathogenic Escherichia coli (ExPEC), which identified a considerable number of genes differentially expressed when the Tat system was disrupted. Among them, a large proportion of flagellar biosynthesis genes showed downregulation, indicating that transcription regulation plays an important role in mediating the motility defects. We further identified three Tat substrate proteins, MdoD, AmiA, and AmiC, that were responsible for the nonmotile phenotype. The Rcs system was deleted in the Δtat, the ΔmdoD, and the ΔamiAΔamiC strains, which restored the motility of ΔmdoD and partially restored the motility of Δtat and ΔamiAΔamiC. The flagella were also observed in all of the ΔtatΔrcsDB, ΔmdoDΔrcsDB, and ΔamiAΔamiCΔrcsDB strains, but not in the Δtat, ΔmdoD, and ΔamiAΔamiC strains, by using transmission electron microscopy. Quantitative reverse transcription-PCR data revealed that the regulons of the Rcs system displayed differential expression in the tat mutant, indicating that the Rcs signaling was activated. Our results suggest that the Rcs system plays an important role in mediating the motility defects of the tat mutant of ExPEC. IMPORTANCE The Tat system is an important protein export system critical for bacterial physiology and pathogenesis. It has been observed for a long time that the Tat system is critical for bacterial motility. However, the underlying mechanism remains unrevealed. In this study, we combine transcriptomics analysis and bacterial genetics, which reveal that transcription regulation plays an important role in mediating the motility defects of the tat mutant of extraintestinal pathogenic Escherichia coli. The Tat substrate proteins responsible for the motility defects are identified. We further show that the Rcs system contributes to the motility suppression. We for the first time reveal the link between the Tat system and bacterial motility, which is important for understanding the physiological functions of the Tat system.
Keywords: Rcs system; extraintestinal pathogenic Escherichia coli; flagella; motility; transcription regulation; twin-arginine translocation system.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
The Tat system and its dependent cell division proteins are critical for virulence of extra-intestinal pathogenic Escherichia coli.Virulence. 2020 Dec;11(1):1279-1292. doi: 10.1080/21505594.2020.1817709. Virulence. 2020. PMID: 32962530 Free PMC article.
-
The role of the bacterial protease Prc in the uropathogenesis of extraintestinal pathogenic Escherichia coli.J Biomed Sci. 2020 Jan 3;27(1):14. doi: 10.1186/s12929-019-0605-y. J Biomed Sci. 2020. PMID: 31900139 Free PMC article.
-
The early mature part of bacterial twin-arginine translocation (Tat) precursor proteins contributes to TatBC receptor binding.J Biol Chem. 2018 May 11;293(19):7281-7299. doi: 10.1074/jbc.RA118.002576. Epub 2018 Mar 28. J Biol Chem. 2018. PMID: 29593092 Free PMC article.
-
Transport and proofreading of proteins by the twin-arginine translocation (Tat) system in bacteria.Biochim Biophys Acta. 2011 Mar;1808(3):876-84. doi: 10.1016/j.bbamem.2010.11.023. Epub 2010 Nov 29. Biochim Biophys Acta. 2011. PMID: 21126506 Review.
-
The twin-arginine protein translocation pathway.Annu Rev Biochem. 2015;84:843-64. doi: 10.1146/annurev-biochem-060614-034251. Epub 2014 Dec 8. Annu Rev Biochem. 2015. PMID: 25494301 Review.
Cited by
-
Functional loss of rffG and rfbB, encoding dTDP-glucose 4,6-dehydratase, alters colony morphology, cell shape, motility and virulence in Salmonella Typhimurium.Front Microbiol. 2025 May 21;16:1572117. doi: 10.3389/fmicb.2025.1572117. eCollection 2025. Front Microbiol. 2025. PMID: 40469742 Free PMC article.
-
Identification of enzymatic functions of osmo-regulated periplasmic glucan biosynthesis proteins from Escherichia coli reveals a novel glycoside hydrolase family.Commun Biol. 2023 Sep 21;6(1):961. doi: 10.1038/s42003-023-05336-6. Commun Biol. 2023. PMID: 37735577 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

