Molecular Evolution of Attachment Glycoprotein (G) and Fusion Protein (F) Genes of Respiratory Syncytial Virus ON1 and BA9 Strains in Xiamen, China
- PMID: 35311585
- PMCID: PMC9045328
- DOI: 10.1128/spectrum.02083-21
Molecular Evolution of Attachment Glycoprotein (G) and Fusion Protein (F) Genes of Respiratory Syncytial Virus ON1 and BA9 Strains in Xiamen, China
Abstract
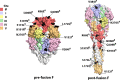
Monitoring viral transmission and analyzing the genetic diversity of a virus are imperative to better understand its evolutionary history and the mechanism driving its evolution and spread. Especially, effective monitoring of key antigenic mutations and immune escape variants caused by these mutations has great scientific importance. Thus, to further understand the molecular evolutionary dynamics of respiratory syncytial virus (RSV) circulating in China, we analyzed nasopharyngeal swab specimens derived from hospitalized children ≤5 years old with acute respiratory tract infections (ARIs) in Xiamen during 2016 to 2019. We found that infants under 6 months of age (52.0%) were the main population with RSV infection. The prevalent pattern "BBAA" of RSV was observed during the epidemic seasons. RSV ON1 and BA9 genotypes were the dominant circulating strains in Xiamen. Interestingly, we observed four Xiamen-specific amino acid substitution combinations in the G protein and several amino acid mutations primarily occurring at antigenic sites Ø and V in the F protein. Our analyses suggest that introduction of new viruses and local evolution are shaping the diversification of RSV strains in Xiamen. This study provides new insights on the evolution and spread of the ON1 and BA9 genotypes at local and global scales. IMPORTANCE Monitoring the amino acid diversity of the RSV G and F genes helps us to find the novel genotypes, key antigenic mutations affecting antigenicity, or neutralizing antibody-resistant variants produced by natural evolution. In this study, we analyzed the molecular evolution of G and F genes from RSV strains circulating in Xiamen, China. These data provide new insights on local and global transmission and could inform the development of control measures for RSV infections.
Keywords: RSV; attachment glycoprotein; fusion protein; hospitalized children; molecular epidemiology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
[Epidemiologic characteristics and the relationship with disease severity of respiratory syncytial virus genotypes from children with lower respiratory tract infection in the southern Zhejiang province].Zhonghua Er Ke Za Zhi. 2015 Jul;53(7):537-41. Zhonghua Er Ke Za Zhi. 2015. PMID: 26310648 Chinese.
-
Respiratory syncytial virus subtype ON1/NA1/BA9 predominates in hospitalized children with lower respiratory tract infections.J Med Virol. 2017 Feb;89(2):213-221. doi: 10.1002/jmv.24619. Epub 2016 Jul 6. J Med Virol. 2017. PMID: 27358012 Free PMC article.
-
Genetic Diversity and Molecular Epidemiology of Circulating Respiratory Syncytial Virus in Central Taiwan, 2008-2017.Viruses. 2021 Dec 24;14(1):32. doi: 10.3390/v14010032. Viruses. 2021. PMID: 35062237 Free PMC article.
-
Global distribution of respiratory syncytial virus A and B infections: a systematic review.Pathog Glob Health. 2022 Oct;116(7):398-409. doi: 10.1080/20477724.2022.2038053. Epub 2022 Feb 14. Pathog Glob Health. 2022. PMID: 35156555 Free PMC article.
-
Clinical and biological consequences of respiratory syncytial virus genetic diversity.Ther Adv Infect Dis. 2022 Oct 8;9:20499361221128091. doi: 10.1177/20499361221128091. eCollection 2022 Jan-Dec. Ther Adv Infect Dis. 2022. PMID: 36225856 Free PMC article. Review.
Cited by
-
AntigenBoost: enhanced mRNA-based antigen expression through rational amino acid substitution.Brief Bioinform. 2024 Sep 23;25(6):bbae468. doi: 10.1093/bib/bbae468. Brief Bioinform. 2024. PMID: 39400114 Free PMC article.
-
Evolutionary trajectory and spread of respiratory syncytial virus group A in neonatal cohorts in Pakistan amidst the COVID-19 pandemic.Sci Rep. 2025 Jul 27;15(1):27329. doi: 10.1038/s41598-025-87332-w. Sci Rep. 2025. PMID: 40717120 Free PMC article.
-
Epidemiology and molecular analyses of respiratory syncytial virus in the 2021-2022 season in northern Italy.Front Microbiol. 2024 Jan 4;14:1327239. doi: 10.3389/fmicb.2023.1327239. eCollection 2023. Front Microbiol. 2024. PMID: 38239726 Free PMC article.
-
Genomic Evolution and Surveillance of Respiratory Syncytial Virus during the 2023-2024 Season.Viruses. 2024 Jul 12;16(7):1122. doi: 10.3390/v16071122. Viruses. 2024. PMID: 39066284 Free PMC article.
-
Whole-Genome Sequencing and Genetic Diversity of Human Respiratory Syncytial Virus in Patients with Influenza-like Illness in Sicily (Italy) from 2017 to 2023.Viruses. 2024 May 26;16(6):851. doi: 10.3390/v16060851. Viruses. 2024. PMID: 38932144 Free PMC article.
References
-
- Shi T, McAllister DA, O’Brien KL, Simoes EAF, Madhi SA, Gessner BD, Polack FP, Balsells E, Acacio S, Aguayo C, Alassani I, Ali A, Antonio M, Awasthi S, Awori JO, Azziz-Baumgartner E, Baggett HC, Baillie VL, Balmaseda A, Barahona A, Basnet S, Bassat Q, Basualdo W, Bigogo G, Bont L, Breiman RF, Brooks WA, Broor S, Bruce N, Bruden D, Buchy P, Campbell S, Carosone-Link P, Chadha M, Chipeta J, Chou M, Clara W, Cohen C, de Cuellar E, Dang DA, Dash-Yandag B, Deloria-Knoll M, Dherani M, Eap T, Ebruke BE, Echavarria M, Emediato CCDL, Fasce RA, Feikin DR, Feng LZ, Gentile A, Gordon A, Goswami D, Goyet S, Groome M, Halasa N, Hirve S, Homaira N, Howie SRC, Jara J, Jroundi I, Kartasasmita CB, Khuri-Bulos N, Kotloff KL, Krishnan A, Libster R, Lopez O, Lucero MG, Lucion F, Lupisan SP, Marcone DN, McCracken JP, Mejia M, Moisi JC, Montgomery JM, Moore DP, Moraleda C, Moyes J, Munywoki P, Mutyara K, Nicol MP, Nokes DJ, Nymadawa P, da Costa Oliveira MT, Oshitani H, Pandey N, Paranhos-Baccalà G, Phillips LN, Picot VS, Rahman M, Rakoto-Andrianarivelo M, Rasmussen ZA, Rath BA, Robinson A, Romero C, Russomando G, Salimi V, Sawatwong P, Scheltema N, Schweiger B, Scott JAG, Seidenberg P, Shen K, Singleton R, Sotomayor V, Strand TA, Sutanto A, Sylla M, Tapia MD, Thamthitiwat S, Thomas ED, Tokarz R, Turner C, Venter M, Waicharoen S, Wang J, Watthanaworawit W, Yoshida LM, Yu H, Zar HJ, Campbell H, Nair H, RSV Global Epidemiology Network. 2017. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet 390:946–958. doi:10.1016/S0140-6736(17)30938-8. - DOI - PMC - PubMed
-
- Baek YH, Choi EH, Song MS, Pascua PN, Kwon HI, Park SJ, Lee JH, Woo SI, Ahn BH, Han HS, Hahn YS, Shin KS, Jang HL, Kim SY, Choi YK. 2012. Prevalence and genetic characterization of respiratory syncytial virus (RSV) in hospitalized children in Korea. Arch Virol 157:1039–1050. doi:10.1007/s00705-012-1267-1. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

