Enemy or ally: a genomic approach to elucidate the lifestyle of Phyllosticta citrichinaensis
- PMID: 35311955
- PMCID: PMC9073689
- DOI: 10.1093/g3journal/jkac061
Enemy or ally: a genomic approach to elucidate the lifestyle of Phyllosticta citrichinaensis
Abstract
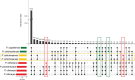
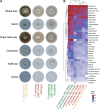
Members of the fungal genus Phyllosticta can colonize a variety of plant hosts, including several Citrus species such as Citrus sinensis (orange), Citrus limon (lemon), and Citrus maxima (pomelo). Some Phyllosticta species have the capacity to cause disease, such as Citrus Black Spot, while others have only been observed as endophytes. Thus far, genomic differences underlying lifestyle adaptations of Phyllosticta species have not yet been studied. Furthermore, the lifestyle of Phyllosticta citrichinaensis is ambiguous, as it has been described as a weak pathogen but Koch's postulates may not have been established and the presence of this species was never reported to cause any crop or economic losses. Here, we examined the genomic differences between pathogenic and endophytic Phyllosticta spp. colonizing Citrus and specifically aimed to elucidate the lifestyle of Phyllosticta citrichinaensis. We found several genomic differences between species of different lifestyles, including groups of genes that were only present in pathogens or endophytes. We also observed that species, based on their carbohydrate active enzymes, group independent of their phylogenetic association, and this clustering correlated with trophy prediction. Phyllosticta citrichinaensis shows an intermediate lifestyle, sharing genomic and phenotypic attributes of both pathogens and endophytes. We thus present the first genomic comparison of multiple citrus-colonizing pathogens and endophytes of the genus Phyllosticta, and therefore provide the basis for further comparative studies into the lifestyle adaptations within this genus.
Keywords: CAZymes; citrus; endophyte; fungal plant pathogens; genomics; lifestyle adaptations; pathogen.
© The Author(s) 2022. Published by Oxford University Press on behalf of Genetics Society of America.
Conflict of interest statement
None declared.
Figures



References
-
- Arafat K. A novel isolate of Phyllosticta capitalensis causes black spot disease on guava fruit in Egypt. Asian J Plant Pathol. 2018;12(1):27–37.
-
- Buijs VA, Zuijdgeest XCL, Groenewald JZ, Crous PW, de Vries RP. Carbon utilization and growth-inhibition of citrus-colonizing Phyllosticta species. Fungal Biol. 2021;125(10):815–825. - PubMed
-
- Chiapello H, Mallet L, Guérin C, Aguileta G, Amselem J, Kroj T, Ortega-Abboud E, Lebrun M-H, Henrissat B, Gendrault A, et al. Deciphering genome content and evolutionary relationships of isolates from the fungus Magnaporthe oryzae attacking different host plants. Genome Biol Evol. 2015;7(10):2896–2912. - PMC - PubMed
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous
