Bacterial community structure and metabolic potential in microbialite-forming mats from South Australian saline lakes
- PMID: 35312212
- PMCID: PMC9311741
- DOI: 10.1111/gbi.12489
Bacterial community structure and metabolic potential in microbialite-forming mats from South Australian saline lakes
Abstract
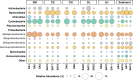
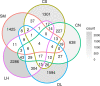
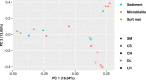
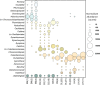
Microbialites are sedimentary rocks created in association with benthic microorganisms. While they harbour complex microbial communities, Cyanobacteria perform critical roles in sediment stabilisation and accretion. Microbialites have been described from permanent and ephemeral saline lakes in South Australia; however, the microbial communities that generate and inhabit these biogeological structures have not been studied in detail. To address this knowledge gap, we investigated the composition, diversity and metabolic potential of bacterial communities from different microbialite-forming mats and surrounding sediments in five South Australian saline coastal lakes using 16S rRNA gene sequencing and predictive metagenome analyses. While Proteobacteria and Bacteroidetes were the dominant phyla recovered from the mats and sediments, Cyanobacteria were significantly more abundant in the mat samples. Interestingly, at lower taxonomic levels, the mat communities were vastly different across the five lakes. Comparative analysis of putative mat and sediment metagenomes via PICRUSt2 revealed important metabolic pathways driving the process of carbonate precipitation, including cyanobacterial oxygenic photosynthesis, ureolysis and nitrogen fixation. These pathways were highly conserved across the five examined lakes, although they appeared to be performed by distinct groups of bacterial taxa found in each lake. Stress response, quorum sensing and circadian clock were other important pathways predicted by the in silico metagenome analysis. The enrichment of CRISPR/Cas and phage shock associated genes in these cyanobacteria-rich communities suggests that they may be under selective pressure from viral infection. Together, these results highlight that a very stable ecosystem function is maintained by distinctly different communities in microbialite-forming mats in the five South Australian lakes and reinforce the concept that 'who' is in the community is not as critical as their net metabolic capacity.
Keywords: 16S rRNA gene amplicon sequencing; bacterial biodiversity; cyanobacteria; microbial mats; microbialites; saline and hypersaline lakes.
© 2022 The Authors. Geobiology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures

Similar articles
-
Unexpected Abundance and Diversity of Phototrophs in Mats from Morphologically Variable Microbialites in Great Salt Lake, Utah.Appl Environ Microbiol. 2020 May 5;86(10):e00165-20. doi: 10.1128/AEM.00165-20. Print 2020 May 5. Appl Environ Microbiol. 2020. PMID: 32198176 Free PMC article.
-
Carbon fixation and rhodopsin systems in microbial mats from hypersaline lakes Brava and Tebenquiche, Salar de Atacama, Chile.PLoS One. 2021 Feb 9;16(2):e0246656. doi: 10.1371/journal.pone.0246656. eCollection 2021. PLoS One. 2021. PMID: 33561170 Free PMC article.
-
Prokaryotic diversity and biogeochemical characteristics of benthic microbial ecosystems at La Brava, a hypersaline lake at Salar de Atacama, Chile.PLoS One. 2017 Nov 15;12(11):e0186867. doi: 10.1371/journal.pone.0186867. eCollection 2017. PLoS One. 2017. PMID: 29140980 Free PMC article.
-
Bacterial Diversity in Microbial Mats and Sediments from the Atacama Desert.Microb Ecol. 2016 Jan;71(1):44-56. doi: 10.1007/s00248-015-0649-9. Epub 2015 Jul 30. Microb Ecol. 2016. PMID: 26224164
-
Microbialite Accretion and Growth: Lessons from Shark Bay and the Bahamas.Ann Rev Mar Sci. 2024 Jan 17;16:487-511. doi: 10.1146/annurev-marine-021423-124637. Ann Rev Mar Sci. 2024. PMID: 38231736 Review.
Cited by
-
Sea spray allows for the growth of subaerial microbialites at the driest desert on Earth.Sci Rep. 2024 Aug 28;14(1):19915. doi: 10.1038/s41598-024-70447-x. Sci Rep. 2024. PMID: 39198637 Free PMC article.
-
Compositional Changes in Sediment Microbiota Are Associated with Seasonal Variation of the Water Column in High-Altitude Hyperarid Andean Lake Systems.Microbiol Spectr. 2023 Jun 15;11(3):e0520022. doi: 10.1128/spectrum.05200-22. Epub 2023 Apr 27. Microbiol Spectr. 2023. PMID: 37102964 Free PMC article.
-
Diversity of sulfur cycling halophiles within the Salton Sea, California's largest lake.BMC Microbiol. 2025 Mar 6;25(1):120. doi: 10.1186/s12866-025-03839-2. BMC Microbiol. 2025. PMID: 40045185 Free PMC article.
-
Microbial Community Shifts and Nitrogen Utilization in Peritidal Microbialites: The Role of Salinity and pH in Microbially Induced Carbonate Precipitation.Microb Ecol. 2025 Apr 22;88(1):31. doi: 10.1007/s00248-025-02532-1. Microb Ecol. 2025. PMID: 40259028 Free PMC article.
References
-
- Águila, B. , Alcántara‐Hernández, R. J. , Montejano, G. , López‐Martínez, R. , Falcón, L. I. , & Becerra‐Absalón, I. (2021). Cyanobacteria in microbialites of Alchichica Crater Lake: A polyphasic characterization. European Journal of Phycology, 56(4), 428–443, 10.1080/09670262.2020.1853815 - DOI
-
- Babilonia, J. , Conesa, A. , Casaburi, G. , Pereira, C. , Louyakis, A. S. , Reid, R. P. , & Foster, J. S. (2018). Comparative metagenomics provides insight into the ecosystem functioning of the shark bay stromatolites, Western Australia. Frontiers in Microbiology, 9(1359), 10.3389/fmicb.2018.01359 - DOI - PMC - PubMed
-
- Baumgartner, L. K. , Reid, R. P. , Dupraz, C. , Decho, A. W. , Buckley, D. H. , Spear, J. R. , Przekop, K. M. , & Visscher, P. T. (2006). Sulfate reducing bacteria in microbial mats: Changing paradigms, new discoveries. Sedimentary Geology, 185(3), 131–145. 10.1016/j.sedgeo.2005.12.008 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

