Single-Molecule Sensor for High-Confidence Detection of miRNA
- PMID: 35312280
- PMCID: PMC9112324
- DOI: 10.1021/acssensors.1c02748
Single-Molecule Sensor for High-Confidence Detection of miRNA
Abstract
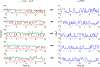
MicroRNAs (miRNAs) play a crucial role in regulating gene expression and have been linked to many diseases. Therefore, sensitive and accurate detection of disease-linked miRNAs is vital to the emerging revolution in early diagnosis of diseases. While the detection of miRNAs is a challenge due to their intrinsic properties such as small size, high sequence similarity among miRNAs and low abundance in biological fluids, the majority of miRNA-detection strategies involve either target/signal amplification or involve complex sensing designs. In this study, we have developed and tested a DNA-based fluorescence resonance energy transfer (FRET) sensor that enables ultrasensitive detection of a miRNA biomarker (miRNA-342-3p) expressed by triple-negative breast cancer (TNBC) cells. The sensor shows a relatively low FRET state in the absence of a target but it undergoes continuous FRET transitions between low- and high-FRET states in the presence of the target. The sensor is highly specific, has a detection limit down to low femtomolar (fM) without having to amplify the target, and has a large dynamic range (3 orders of magnitude) extending to 300 000 fM. Using this strategy, we demonstrated that the sensor allows detection of miRNA-342-3p in the miRNA-extracts from cancer cell lines and TNBC patient-derived xenografts. Given the simple-to-design hybridization-based detection, the sensing platform developed here can be used to detect a wide range of miRNAs enabling early diagnosis and screening of other genetic disorders.
Keywords: biomarkers; fluorescence resonance energy transfer (FRET); high-confidence; miRNA; single-molecule; triple negative breast cancer (TNBC).
Figures
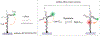

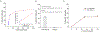
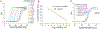
References
-
- Chen J; Chen Z; Huang J; Chen F; Ye W; Ding G; Wang X Bioinformatics Identification of Dysregulated MicroRNAs in Triple Negative Breast Cancer Based on MicroRNA Expression Profiling. Oncol. Lett 2017. 10.3892/ol.2017.7707. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

