A GCDGC-specific DNA (cytosine-5) methyltransferase that methylates the GCWGC sequence on both strands and the GCSGC sequence on one strand
- PMID: 35312710
- PMCID: PMC8936443
- DOI: 10.1371/journal.pone.0265225
A GCDGC-specific DNA (cytosine-5) methyltransferase that methylates the GCWGC sequence on both strands and the GCSGC sequence on one strand
Abstract
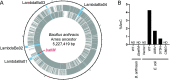
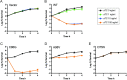
5-Methylcytosine is one of the major epigenetic marks of DNA in living organisms. Some bacterial species possess DNA methyltransferases that modify cytosines on both strands to produce fully-methylated sites or on either strand to produce hemi-methylated sites. In this study, we characterized a DNA methyltransferase that produces two sequences with different methylation patterns: one methylated on both strands and another on one strand. M.BatI is the orphan DNA methyltransferase of Bacillus anthracis coded in one of the prophages on the chromosome. Analysis of M.BatI modified DNA by bisulfite sequencing revealed that the enzyme methylates the first cytosine in sequences of 5'-GCAGC-3', 5'-GCTGC-3', and 5'-GCGGC-3', but not of 5'-GCCGC-3'. This resulted in the production of fully-methylated 5'-GCWGC-3' and hemi-methylated 5'-GCSGC-3'. M.BatI also showed toxicity when expressed in E. coli, which was caused by a mechanism other than DNA modification activity. Homologs of M.BatI were found in other Bacillus species on different prophage like regions, suggesting the spread of the gene by several different phages. The discovery of the DNA methyltransferase with unique modification target specificity suggested unrevealed diversity of target sequences of bacterial cytosine DNA methyltransferase.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Putative DNA modification methylase DR_C0020 of Deinococcus radiodurans is an atypical SAM dependent C-5 cytosine DNA methylase.Biochim Biophys Acta Gen Subj. 2017 Mar;1861(3):593-602. doi: 10.1016/j.bbagen.2016.12.025. Epub 2016 Dec 27. Biochim Biophys Acta Gen Subj. 2017. PMID: 28038990
-
The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites.J Biol Chem. 2004 Nov 12;279(46):48350-9. doi: 10.1074/jbc.M403427200. Epub 2004 Aug 31. J Biol Chem. 2004. PMID: 15339928
-
Absence in Bacillus subtilis and Staphylococcus aureus of the sequence-specific deoxyribonucleic acid methylation that is conferred in Escherichia coli K-12 by the dam and dcm enzymes.J Bacteriol. 1981 Jul;147(1):259-61. doi: 10.1128/jb.147.1.259-261.1981. J Bacteriol. 1981. PMID: 6263867 Free PMC article.
-
DNA Methyltransferases in Mammalian Oocytes.Results Probl Cell Differ. 2017;63:211-222. doi: 10.1007/978-3-319-60855-6_10. Results Probl Cell Differ. 2017. PMID: 28779320 Review.
-
Mammalian DNA (cytosine-5) methyltransferases and their expression.Clin Immunol. 2003 Oct;109(1):6-16. doi: 10.1016/s1521-6616(03)00204-3. Clin Immunol. 2003. PMID: 14585271 Review.
Cited by
-
Structural analysis of the BisI family of modification dependent restriction endonucleases.Nucleic Acids Res. 2024 Aug 27;52(15):9103-9118. doi: 10.1093/nar/gkae634. Nucleic Acids Res. 2024. PMID: 39041409 Free PMC article.
-
Correcting modification-mediated errors in nanopore sequencing by nucleotide demodification and reference-based correction.Commun Biol. 2023 Nov 29;6(1):1215. doi: 10.1038/s42003-023-05605-4. Commun Biol. 2023. PMID: 38030695 Free PMC article.
-
DNA methylases for site-selective inhibition of type IIS restriction enzyme activity.Appl Microbiol Biotechnol. 2024 Jan 25;108(1):174. doi: 10.1007/s00253-024-13015-7. Appl Microbiol Biotechnol. 2024. PMID: 38270650 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

