Detection of SARS-CoV-2 and the L452R spike mutation using reverse transcription loop-mediated isothermal amplification plus bioluminescent assay in real-time (RT-LAMP-BART)
- PMID: 35312732
- PMCID: PMC8936440
- DOI: 10.1371/journal.pone.0265748
Detection of SARS-CoV-2 and the L452R spike mutation using reverse transcription loop-mediated isothermal amplification plus bioluminescent assay in real-time (RT-LAMP-BART)
Abstract
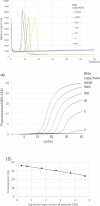
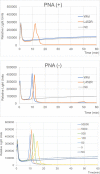
The new coronavirus infection (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can be fatal, and several variants of SARS-CoV-2 with mutations of the receptor-binding domain (RBD) have increased avidity for human cell receptors. A single missense mutation of U to G at nucleotide position 1355 (U1355G) in the spike (S) gene changes leucine to arginine (L452R) in the spike protein. This mutation has been observed in the India and California strains (B.1.617 and B.1.427/B.1.429, respectively). Control of COVID-19 requires rapid and reliable detection of SARS-CoV-2. Therefore, we established a reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay plus a bioluminescent assay in real-time (BART) to detect SARS-CoV-2 and the L452R spike mutation. The specificity and sensitivity of the RT-LAMP-BART assay was evaluated using synthetic RNAs including target sequences and RNA-spiked clinical nasopharyngeal and saliva specimens as well as reference strains representing five viral and four bacterial pathogens. The novel RT-LAMP-BART assay to detect SARS-CoV-2 was highly specific compared to the conventional real-time RT-PCR. Within 25 min, the RT-LAMP-BART assay detected 80 copies of the target gene in a sample, whereas the conventional real-time RT-PCR method detected 5 copies per reaction within 130 min. Using RNA-spiked specimens, the sensitivity of the RT-LAMP-BART assay was slightly attenuated compared to purified RNA as a template. The results were identical to those of the conventional real-time RT-PCR method. Furthermore, using a peptide nucleic acid (PNA) probe, the RT-LAMP-BART method correctly identified the L452R spike mutation. This is the first report describes RT-LAMP-BART as a simple, inexpensive, rapid, and useful assay for detection of SARS-CoV-2, its variants of concern, and for screening of COVID-19.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: LT is an employee of Erba Molecular. NP is an employee of 3M Company. This does not alter our adherence to PLOS ONE policies on sharing data and materials. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.The statements do not alter the authors’ adherence to PLOS ONE policies on sharing data and materials.
Figures
Similar articles
-
A colorimetric reverse-transcription loop-mediated isothermal amplification method targeting the L452R mutation to detect the Delta variant of SARS-CoV-2.Sci Rep. 2024 Sep 20;14(1):21961. doi: 10.1038/s41598-024-72417-9. Sci Rep. 2024. PMID: 39304686 Free PMC article.
-
A New Method to Detect Variants of SARS-CoV-2 Using Reverse Transcription Loop-Mediated Isothermal Amplification Combined with a Bioluminescent Assay in Real Time (RT-LAMP-BART).Int J Mol Sci. 2023 Jun 27;24(13):10698. doi: 10.3390/ijms241310698. Int J Mol Sci. 2023. PMID: 37445876 Free PMC article.
-
A Novel Approach to the Bioluminescent Detection of the SARS-CoV-2 ORF1ab Gene by Coupling Isothermal RNA Reverse Transcription Amplification with a Digital PCR Approach.Int J Mol Sci. 2021 Jan 20;22(3):1017. doi: 10.3390/ijms22031017. Int J Mol Sci. 2021. PMID: 33498408 Free PMC article.
-
A loop-mediated isothermal amplification-enabled analytical assay for the detection of SARS-CoV-2: A review.Front Cell Infect Microbiol. 2022 Dec 23;12:1068015. doi: 10.3389/fcimb.2022.1068015. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36619749 Free PMC article. Review.
-
Nucleic Acid Testing of SARS-CoV-2.Int J Mol Sci. 2021 Jun 7;22(11):6150. doi: 10.3390/ijms22116150. Int J Mol Sci. 2021. PMID: 34200331 Free PMC article. Review.
Cited by
-
Rapid assays of SARS-CoV-2 virus and noble biosensors by nanomaterials.Nano Converg. 2024 Jan 8;11(1):2. doi: 10.1186/s40580-023-00408-z. Nano Converg. 2024. PMID: 38190075 Free PMC article. Review.
-
Multiplexed Reverse Transcription Loop-Mediated Isothermal Amplification Coupled with a Nucleic Acid-Based Lateral Flow Dipstick as a Rapid Diagnostic Method to Detect SARS-CoV-2.Microorganisms. 2023 May 7;11(5):1233. doi: 10.3390/microorganisms11051233. Microorganisms. 2023. PMID: 37317207 Free PMC article.
-
Versatility of reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) from diagnosis of early pathological infection to mutation detection in organisms.Mol Biol Rep. 2024 Jan 25;51(1):211. doi: 10.1007/s11033-023-09110-z. Mol Biol Rep. 2024. PMID: 38270670 Review.
-
A colorimetric reverse-transcription loop-mediated isothermal amplification method targeting the L452R mutation to detect the Delta variant of SARS-CoV-2.Sci Rep. 2024 Sep 20;14(1):21961. doi: 10.1038/s41598-024-72417-9. Sci Rep. 2024. PMID: 39304686 Free PMC article.
-
Fluorometric Detection of SARS-CoV-2 Single-Nucleotide Variant L452R Using Ligation-Based Isothermal Gene Amplification.Bioengineering (Basel). 2023 Sep 23;10(10):1116. doi: 10.3390/bioengineering10101116. Bioengineering (Basel). 2023. PMID: 37892846 Free PMC article.
References
-
- Larsen HD, Fonager J, Lomholt FK, Dalby T, Benedetti G, Kristensen B, et al.. Preliminary report of an outbreak of SARS-CoV-2 in mink and mink farmers associated with community spread, Denmark, June to November 2020. Euro Surveill. 2021; 26(5). doi: 10.2807/1560-7917.ES.2021.26.5.210009 . - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

