DRP1 contributes to head and neck cancer progression and induces glycolysis through modulated FOXM1/MMP12 axis
- PMID: 35313071
- PMCID: PMC9251862
- DOI: 10.1002/1878-0261.13212
DRP1 contributes to head and neck cancer progression and induces glycolysis through modulated FOXM1/MMP12 axis
Abstract
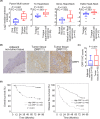
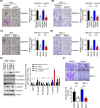
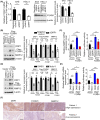
Abnormal DRP1 expression has been identified in a variety of human cancers. However, the prognostic potential and mechanistic role of DRP1 in head and neck cancer (HNC) are currently poorly understood. Here, we demonstrated a significant upregulation of DRP1 in HNC tissues, and that DRP1 expression correlates with poor survival of HNC patients. Diminished DRP1 expression suppressed tumor growth and metastasis in both in vitro and in vivo models. DRP1 expression was positively correlated with FOXM1 and MMP12 expression in HNC patient samples, suggesting pathological relevance in the context of HNC development. Moreover, DRP1 depletion affected aerobic glycolysis through the downregulation of glycolytic genes, and overexpression of MMP12 in DRP1-depleted cells could help restore glucose consumption and lactate production. Using ChIP-qPCR, we showed that DRP1 modulates FOXM1 expression, which can enhance MMP12 transcription by binding to its promoter. We also showed that miR-575 could target 3'UTR of DRP1 mRNA and suppress DRP1 expression. Collectively, our study provides mechanistic insights into the role of DRP1 in HNC and highlights the potential of targeting the miR-575/DRP1/FOXM1/MMP12 axis as a novel therapy for the prevention of HNC progression.
Keywords: DRP1; FOXM1; MMP12; glycolysis; head and neck cancer; miR-575.
© 2022 The Authors. Molecular Oncology published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
-
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. - PubMed
-
- Chow LQM. Head and neck cancer. N Engl J Med. 2020;382:60–72. - PubMed
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. Cancer J Clin. 2018;68:7–30. - PubMed
-
- Cancer Registry Annual Report, 2017. Taiwan.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

