Chemical modifications to mRNA nucleobases impact translation elongation and termination
- PMID: 35313212
- PMCID: PMC9373004
- DOI: 10.1016/j.bpc.2022.106780
Chemical modifications to mRNA nucleobases impact translation elongation and termination
Abstract
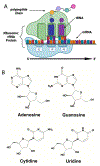
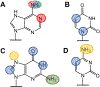
Messenger RNAs (mRNAs) serve as blueprints for protein synthesis by the molecular machine the ribosome. The ribosome relies on hydrogen bonding interactions between adaptor aminoacyl-transfer RNA molecules and mRNAs to ensure the rapid and faithful translation of the genetic code into protein. There is a growing body of evidence suggesting that chemical modifications to mRNA nucleosides impact the speed and accuracy of protein synthesis by the ribosome. Modulations in translation rates have downstream effects beyond protein production, influencing protein folding and mRNA stability. Given the prevalence of such modifications in mRNA coding regions, it is imperative to understand the consequences of individual modifications on translation. In this review we present the current state of our knowledge regarding how individual mRNA modifications influence ribosome function. Our comprehensive comparison of the impacts of 16 different mRNA modifications on translation reveals that most modifications can alter the elongation step in the protein synthesis pathway. Additionally, we discuss the context dependence of these effects, highlighting the necessity of further study to uncover the rules that govern how any given chemical modification in an mRNA codon is read by the ribosome.
Keywords: Kinetics; RNA modification; Translation; mRNA modification.
Copyright © 2022. Published by Elsevier B.V.
Conflict of interest statement
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
Pseudouridinylation of mRNA coding sequences alters translation.Proc Natl Acad Sci U S A. 2019 Nov 12;116(46):23068-23074. doi: 10.1073/pnas.1821754116. Epub 2019 Oct 31. Proc Natl Acad Sci U S A. 2019. PMID: 31672910 Free PMC article.
-
Isolation of ribosome bound nascent polypeptides in vitro to identify translational pause sites along mRNA.J Vis Exp. 2012 Jul 6;(65):4026. doi: 10.3791/4026. J Vis Exp. 2012. PMID: 22806127 Free PMC article.
-
Translation elongation: measurements and applications.RNA Biol. 2025 Dec;22(1):1-10. doi: 10.1080/15476286.2025.2504727. Epub 2025 May 16. RNA Biol. 2025. PMID: 40377059 Free PMC article. Review.
-
Codon optimality-mediated mRNA degradation: Linking translational elongation to mRNA stability.Mol Cell. 2022 Apr 21;82(8):1467-1476. doi: 10.1016/j.molcel.2022.03.032. Mol Cell. 2022. PMID: 35452615 Free PMC article. Review.
-
The ribosome in action: Tuning of translational efficiency and protein folding.Protein Sci. 2016 Aug;25(8):1390-406. doi: 10.1002/pro.2950. Epub 2016 Jun 8. Protein Sci. 2016. PMID: 27198711 Free PMC article. Review.
Cited by
-
mRNA vaccines in the prevention and treatment of diseases.MedComm (2020). 2022 Aug 25;3(3):e167. doi: 10.1002/mco2.167. eCollection 2022 Sep. MedComm (2020). 2022. PMID: 36033422 Free PMC article. Review.
-
β-Aminobutyric acid promotes stress tolerance, physiological adjustments, as well as broad epigenetic changes at DNA and RNA nucleobases in field elms (Ulmus minor).BMC Plant Biol. 2024 Aug 15;24(1):779. doi: 10.1186/s12870-024-05425-6. BMC Plant Biol. 2024. PMID: 39148013 Free PMC article.
-
mRNA Regulation by RNA Modifications.Annu Rev Biochem. 2023 Jun 20;92:175-198. doi: 10.1146/annurev-biochem-052521-035949. Epub 2023 Apr 5. Annu Rev Biochem. 2023. PMID: 37018844 Free PMC article. Review.
-
The Potential of RNA Therapeutics in Treating Cardiovascular Disease.Drugs. 2025 May;85(5):659-676. doi: 10.1007/s40265-025-02173-1. Epub 2025 Apr 2. Drugs. 2025. PMID: 40175855 Review.
-
Probing enzyme-dependent pseudouridylation using direct RNA sequencing to assess epitranscriptome plasticity in a neuronal cell line.Cell Syst. 2025 Apr 16;16(4):101238. doi: 10.1016/j.cels.2025.101238. Epub 2025 Mar 20. Cell Syst. 2025. PMID: 40118059
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

