Patient-specific deep learning model to enhance 4D-CBCT image for radiomics analysis
- PMID: 35313293
- PMCID: PMC9066277
- DOI: 10.1088/1361-6560/ac5f6e
Patient-specific deep learning model to enhance 4D-CBCT image for radiomics analysis
Abstract
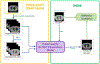
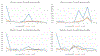
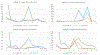
Objective.4D-CBCT provides phase-resolved images valuable for radiomics analysis for outcome prediction throughout treatment courses. However, 4D-CBCT suffers from streak artifacts caused by under-sampling, which severely degrades the accuracy of radiomic features. Previously we developed group-patient-trained deep learning methods to enhance the 4D-CBCT quality for radiomics analysis, which was not optimized for individual patients. In this study, a patient-specific model was developed to further improve the accuracy of 4D-CBCT based radiomics analysis for individual patients.Approach.This patient-specific model was trained with intra-patient data. Specifically, patient planning 4D-CT was augmented through image translation, rotation, and deformation to generate 305 CT volumes from 10 volumes to simulate possible patient positions during the onboard image acquisition. 72 projections were simulated from 4D-CT for each phase and were used to reconstruct 4D-CBCT using FDK back-projection algorithm. The patient-specific model was trained using these 305 paired sets of patient-specific 4D-CT and 4D-CBCT data to enhance the 4D-CBCT image to match with 4D-CT images as ground truth. For model testing, 4D-CBCT were simulated from a separate set of 4D-CT scan images acquired from the same patient and were then enhanced by this patient-specific model. Radiomics features were then extracted from the testing 4D-CT, 4D-CBCT, and enhanced 4D-CBCT image sets for comparison. The patient-specific model was tested using 4 lung-SBRT patients' data and compared with the performance of the group-based model. The impact of model dimensionality, region of interest (ROI) selection, and loss function on the model accuracy was also investigated.Main results.Compared with a group-based model, the patient-specific training model further improved the accuracy of radiomic features, especially for features with large errors in the group-based model. For example, the 3D whole-body and ROI loss-based patient-specific model reduces the errors of the first-order median feature by 83.67%, the wavelet LLL feature maximum by 91.98%, and the wavelet HLL skewness feature by 15.0% on average for the four patients tested. In addition, the patient-specific models with different dimensionality (2D versus 3D) or loss functions (L1 versus L1 + VGG + GAN) achieved comparable results for improving the radiomics accuracy. Using whole-body or whole-body+ROI L1 loss for the model achieved better results than using the ROI L1 loss alone as the loss function.Significance.This study demonstrated that the patient-specific model is more effective than the group-based model on improving the accuracy of the 4D-CBCT radiomic features analysis, which could potentially improve the precision for outcome prediction in radiotherapy.
Keywords: 4D-CBCT; CBCT; deep learning; generative adversarial network; patient-specific; radiomics; under-sampled projections.
© 2022 Institute of Physics and Engineering in Medicine.
Figures







References
-
- Feldkamp LA, Davis LC and Kress JW 1984. Practical cone-beam algorithm Josa a 1 612–9
-
- Ganeshan B, Panayiotou E, Burnand K, Dizdarevic S and Miles K 2012. Tumour heterogeneity in non-small cell lung carcinoma assessed by CT texture analysis: a potential marker of survival European radiology 22 796–802 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
