Effect of insilico predicted and designed potential siRNAs on inhibition of SARS-CoV-2 in HEK-293 cells
- PMID: 35313445
- PMCID: PMC8925144
- DOI: 10.1016/j.jksus.2022.101965
Effect of insilico predicted and designed potential siRNAs on inhibition of SARS-CoV-2 in HEK-293 cells
Abstract
Objectives: The COVID-19 was identified for the first time from the sea food market, Wuhan city, China in 2019 and the pathogenic organism was identified as SARS-CoV-2. Currently, this virus has spread to 223 countries and territories and known as a serious issue for the global human community. Many vaccines have been developed and used for immunization.
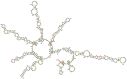
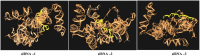
Methods: We have reported the insilico prediction, designing, secondary structure prediction, molecular docking analysis, and in vitro assessment of siRNAs against SARS-CoV-2. The online bioinformatic approach was used for siRNAs selection and designing. The selected siRNAs were evaluated for antiviral efficacy by using Lipofectamine 2000 as delivery agent to HEK-293 cells. The MTT assay was used for cytotoxicity determination. The antiviral efficacy of potential siRNAs was determined based on the Ct value of q-RT-PCR and the data analysis was done by Prism-GraphPad software.
Results: The analyzed data resulted in the selection of only three siRNAs out of twenty-six siRNAs generated by online software. The secondary structure prediction and molecular docking analysis of siRNAs revealed the efficient binding to the target. There was no cellular toxicity observed in the HEK-293 cells at any tested concentrations of siRNAs. The purification of RNA was completed from inoculated cells and subjected to q-RT-PCR. The highest Ct value was observed in siRNA 3 than the others. The results offered valuable evidence and invigorated us to assess the potency of siRNAs by using alone or in combination in other human cells.
Conclusion: The data generated from this study indicates the significance of in silico prediction and narrow down the potential siRNA' against SARS-CoV-2, and molecular docking investigation offered the effective siRNAs binding with the target. Finally, it is concluded that the online bioinformatics approach provided the prediction and selection of siRNAs with better antiviral efficacy. The siRNA-3 was observed to be the best for reduction of viral RNA in cells.
Keywords: COVID-19, The new Coronavirus Disease 2019; Designing; HEK-293 cells; In silico prediction; MFE, Minimum free energy; PCR, Polymerase Chain Reaction; RNAi, RNA interference; SARS-CoV-2; SARS-CoV-2, Severe Acute Respiratory Syndrome Coranavirus-2; WHO, World Health Organization; siRNA, short interfering RNA; siRNAs.
© 2022 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
-
- Alnylam Pharmaceuticals, 2020. Vir and Alnylam Expand Collaboration to Advance RNAi Therapeutics for the Treatment of Coronavirus Infection, Including Covid-19. Available online at: https://investors.alnylam.com/press-release?id= 24656.
-
- Aleem, et al. Stat Pearls Publishing; In Stat Pearls: 2022. Emerging Variants of SARS-CoV-2 And Novel Therapeutics Against Coronavirus (COVID-19) - PubMed
-
- Azoulay E., Fartoukh M., Darmon M., Géri G., Voiriot G., Dupont T., Zafrani L., Girodias L., Labbé V., Dres M., Beurton A., Vieillard-Baron A., Demoule A. Increased mortality in patients with severe SARS-CoV-2 infection admitted within seven days of disease onset. Intensive Care Med. 2020;46(9):1714–1722. - PMC - PubMed
-
- Bellaousov et al., 2013. RNA structure: Web servers for RNA secondary structure prediction and analysis. Nucl. Acids Res., 41(Web Server issue), W471–W474. https://doi.org/10.1093/nar/gkt290. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
